This is a classification scenario where you try to predict a categorical binary target y if the person survived (1) or not (0) from the Titanic.
This example is really short and here just to cover an example of classification as we mainly focused on regression so far.
Most of the supervised learning workflow does not change. You will most likely use classifier estimators from scikit, can also pick a different loss function, and a global metric that is most suited for your use-case.
url = 'https://gist.githubusercontent.com/michhar/2dfd2de0d4f8727f873422c5d959fff5/raw/fa71405126017e6a37bea592440b4bee94bf7b9e/titanic.csv'
First imports
Some may be added along with this practice session
import matplotlib.pyplot as plt
import seaborn as sns
import pandas as pd
import numpy as np
Download the dataset
using shell command wget
!wget -O montitanic.csv $url
pandas.read_csv can also read directly from a URL
pd.set_option("max_rows", 100) # just for showing more lines by default
df = pd.read_csv(url)
df.head(3)
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 3 | Braund, Mr. Owen Harris | male | 22.0 | 1 | 0 | A/5 21171 | 7.2500 | NaN | S |
| 1 | 2 | 1 | 1 | Cumings, Mrs. John Bradley (Florence Briggs Th... | female | 38.0 | 1 | 0 | PC 17599 | 71.2833 | C85 | C |
| 2 | 3 | 1 | 3 | Heikkinen, Miss. Laina | female | 26.0 | 0 | 0 | STON/O2. 3101282 | 7.9250 | NaN | S |
Health checks
Checking the documentation of the dataset
The target
- y = survived indicator (0 No, 1 yes)
The features
- Pclass = passenger class: 1st class, 2nd class, 3rd class
- name = name of the person
- sex
- age
- sibsip = number of siblings/spouses who traveled with the person
- parch = number of parents (children?) who traveled with the person
- ticket = ticket number / identifier
- fare = ticket price in pounds
- cabin = cabin type
- embarked = ferry port / jetty
Checks on the dataset
nb of rows / columns
df.shape
(891, 12)
Checking the columns
df.columns
Index(['PassengerId', 'Survived', 'Pclass', 'Name', 'Sex', 'Age', 'SibSp',
'Parch', 'Ticket', 'Fare', 'Cabin', 'Embarked'],
dtype='object')
Checking the proportion of each values the binary target can take
df.Survived.value_counts() # the target = y, imbalanced/balanced dataset ?
0 549
1 342
Name: Survived, dtype: int64
df.Survived.value_counts().plot(kind="bar")
<AxesSubplot:>
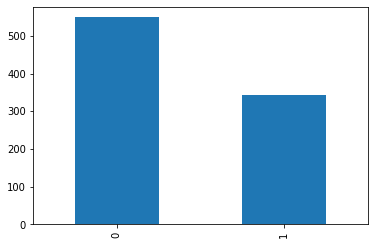
infered types of each column
df.dtypes
PassengerId int64
Survived int64
Pclass int64
Name object
Sex object
Age float64
SibSp int64
Parch int64
Ticket object
Fare float64
Cabin object
Embarked object
dtype: object
Categorical vs Numerical Features
which features are categorical ? numerical ?
- categorical: Sex, Name, titleName, Embarked, Ticket
- ordinal: Pclass
- numerical:
- continuous=Age, Fare
- discrete=SibSp, Parch
The outcome variable y is categorical
categorical_cols = ["Sex", "Embarked", "Pclass", "Cabin", "Ticket"]
numerical_cols = ["Age", "Fare", "SibSp", "Parch"]
Let’s convert categorical columns as is
df[categorical_cols] = df[categorical_cols].astype("category")
df.dtypes
PassengerId int64
Survived int64
Pclass category
Name object
Sex category
Age float64
SibSp int64
Parch int64
Ticket category
Fare float64
Cabin category
Embarked category
dtype: object
len(categorical_cols ) + len(numerical_cols)
9
set(df.columns) - set(categorical_cols + numerical_cols)
{'Name', 'PassengerId', 'Survived'}
number of unique values for each column
df.nunique()
PassengerId 891
Survived 2
Pclass 3
Name 891
Sex 2
Age 88
SibSp 7
Parch 7
Ticket 681
Fare 248
Cabin 147
Embarked 3
dtype: int64
df.drop("Ticket", axis=1, inplace=True)
categorical_cols.remove("Ticket")
an easy-win: df.describe()
df[numerical_cols].describe()
| Age | Fare | SibSp | Parch | |
|---|---|---|---|---|
| count | 714.000000 | 891.000000 | 891.000000 | 891.000000 |
| mean | 29.699118 | 32.204208 | 0.523008 | 0.381594 |
| std | 14.526497 | 49.693429 | 1.102743 | 0.806057 |
| min | 0.420000 | 0.000000 | 0.000000 | 0.000000 |
| 25% | 20.125000 | 7.910400 | 0.000000 | 0.000000 |
| 50% | 28.000000 | 14.454200 | 0.000000 | 0.000000 |
| 75% | 38.000000 | 31.000000 | 1.000000 | 0.000000 |
| max | 80.000000 | 512.329200 | 8.000000 | 6.000000 |
df[categorical_cols].describe()
| Sex | Embarked | Pclass | Cabin | |
|---|---|---|---|---|
| count | 891 | 889 | 891 | 204 |
| unique | 2 | 3 | 3 | 147 |
| top | male | S | 3 | B96 B98 |
| freq | 577 | 644 | 491 | 4 |
Preprocessing (example)
Let’s try to extract the sexe gender of a person based on his name and cross-check with the sex column.
df['titleName'] = df.Name.str.extract("(?i)(mrs|mr|miss)")
print( df.loc[df.Sex == "male", "titleName"].isin(["miss", "mrs"]).any() )
print( df.loc[df.Sex == "female", "titleName"].isin(["mr"]).any() )
False
False
Good thing here, the sex type is matching the particle in the name (Mr = male, Miss and Mrs = female)
df.titleName.isna().mean() *100
7.182940516273851
Though, there is still 7% of missing values from the transformation of the name, let’s further check this.m
We see the particle is always followed by a dot, let’s try to extract it this way then.
df["titleName"] = df.Name.str.extract("([a-zA-Z]+)\.")
df.titleName.value_counts().plot(kind="bar")
<AxesSubplot:>
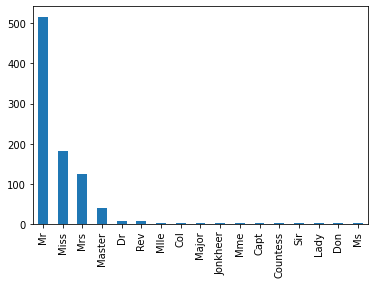
Mr, Miss and Mrs are the most represented titleName.
Let’s check with all the different values’ proportions.
labels = df.titleName.value_counts()
labels
Mr 517
Miss 182
Mrs 125
Master 40
Dr 7
Rev 6
Mlle 2
Col 2
Major 2
Jonkheer 1
Mme 1
Capt 1
Countess 1
Sir 1
Lady 1
Don 1
Ms 1
Name: titleName, dtype: int64
Some labels/titleName are really minorities. Let’s regroup them in “other”.
df.loc[df.titleName.isin(labels[labels<10].index), "titleName"] = "other"
Adding to the categorical columns:
categorical_cols.append('titleName')
Some other Exploratory Data Analysis
ticket prices distribution
df.Fare.min()
0.0
df.Fare.plot(kind="density", xlim=(df.Fare.min(), df.Fare.max()))
<AxesSubplot:ylabel='Density'>
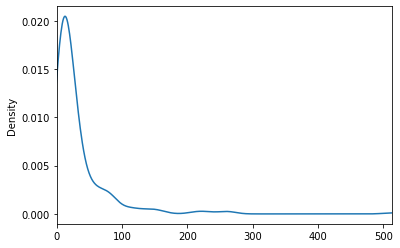
highly right-skewed, who paid so much ?
df.loc[ df.Fare == df.Fare.max()]
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Fare | Cabin | Embarked | titleName | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 258 | 259 | 1 | 1 | Ward, Miss. Anna | female | 35.0 | 0 | 0 | 512.3292 | NaN | C | Miss |
| 679 | 680 | 1 | 1 | Cardeza, Mr. Thomas Drake Martinez | male | 36.0 | 0 | 1 | 512.3292 | B51 B53 B55 | C | Mr |
| 737 | 738 | 1 | 1 | Lesurer, Mr. Gustave J | male | 35.0 | 0 | 0 | 512.3292 | B101 | C | Mr |
who does not pay anything for onboarding ?
df.loc[ df.Fare == df.Fare.min()]
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Fare | Cabin | Embarked | titleName | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 179 | 180 | 0 | 3 | Leonard, Mr. Lionel | male | 36.0 | 0 | 0 | 0.0 | NaN | S | Mr |
| 263 | 264 | 0 | 1 | Harrison, Mr. William | male | 40.0 | 0 | 0 | 0.0 | B94 | S | Mr |
| 271 | 272 | 1 | 3 | Tornquist, Mr. William Henry | male | 25.0 | 0 | 0 | 0.0 | NaN | S | Mr |
| 277 | 278 | 0 | 2 | Parkes, Mr. Francis "Frank" | male | NaN | 0 | 0 | 0.0 | NaN | S | Mr |
| 302 | 303 | 0 | 3 | Johnson, Mr. William Cahoone Jr | male | 19.0 | 0 | 0 | 0.0 | NaN | S | Mr |
| 413 | 414 | 0 | 2 | Cunningham, Mr. Alfred Fleming | male | NaN | 0 | 0 | 0.0 | NaN | S | Mr |
| 466 | 467 | 0 | 2 | Campbell, Mr. William | male | NaN | 0 | 0 | 0.0 | NaN | S | Mr |
| 481 | 482 | 0 | 2 | Frost, Mr. Anthony Wood "Archie" | male | NaN | 0 | 0 | 0.0 | NaN | S | Mr |
| 597 | 598 | 0 | 3 | Johnson, Mr. Alfred | male | 49.0 | 0 | 0 | 0.0 | NaN | S | Mr |
| 633 | 634 | 0 | 1 | Parr, Mr. William Henry Marsh | male | NaN | 0 | 0 | 0.0 | NaN | S | Mr |
| 674 | 675 | 0 | 2 | Watson, Mr. Ennis Hastings | male | NaN | 0 | 0 | 0.0 | NaN | S | Mr |
| 732 | 733 | 0 | 2 | Knight, Mr. Robert J | male | NaN | 0 | 0 | 0.0 | NaN | S | Mr |
| 806 | 807 | 0 | 1 | Andrews, Mr. Thomas Jr | male | 39.0 | 0 | 0 | 0.0 | A36 | S | Mr |
| 815 | 816 | 0 | 1 | Fry, Mr. Richard | male | NaN | 0 | 0 | 0.0 | B102 | S | Mr |
| 822 | 823 | 0 | 1 | Reuchlin, Jonkheer. John George | male | 38.0 | 0 | 0 | 0.0 | NaN | S | other |
Proportion of each value each categorical feature may take
categorical_cols.remove('Cabin') # we will explain why later on
n = len(categorical_cols)
fig, axes = plt.subplots(1, n, figsize=(15, 4), sharey=True)
for i, colname in enumerate(categorical_cols):
sns.countplot(x=colname, data=df, ax=axes[i])
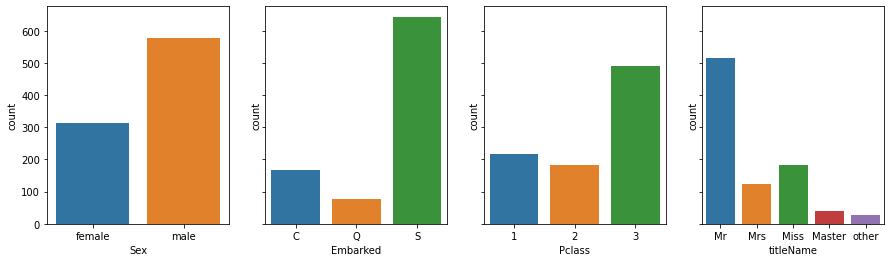
and with respect to the target
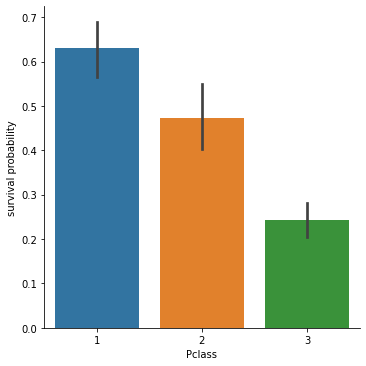
From the next plot we can see that the Survival probability is linked with the membership to a Pclass value.
sns.catplot(x="Pclass", y="Survived", kind="bar", data=df).set_ylabels("survival probability")
<seaborn.axisgrid.FacetGrid at 0x178b37220>
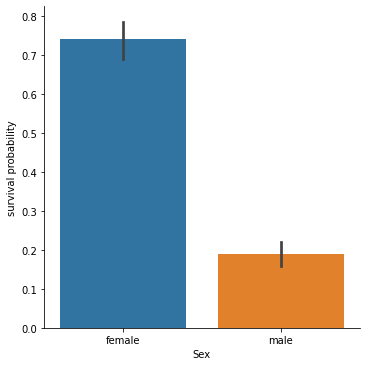
But also to the sex category of a person.
sns.catplot(x="Sex", y="Survived", kind="bar", data=df).set_ylabels("survival probability")
<seaborn.axisgrid.FacetGrid at 0x178b0e040>
We may want to further quantify this relationship using statistical tests.
Let’s keep our investigations going on.
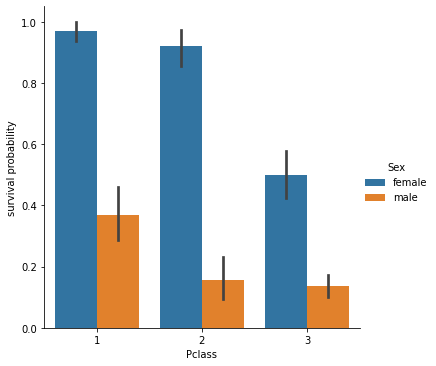
Trying to check the influence of both features on the survival problability (does a female in 3rd class had more chance to survive to the Titanic compared to a male on 1st class ?)
sns.catplot(x="Pclass", y="Survived", hue="Sex", kind="bar", data=df).set_ylabels("survival probability")
<seaborn.axisgrid.FacetGrid at 0x178bd0fd0>
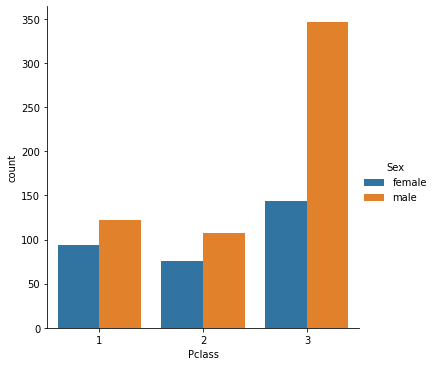
And let’s also check the number of people constituing each of those subgroups.
sns.catplot(x="Pclass",hue="Sex", kind="count", data=df)
<seaborn.axisgrid.FacetGrid at 0x178c499d0>
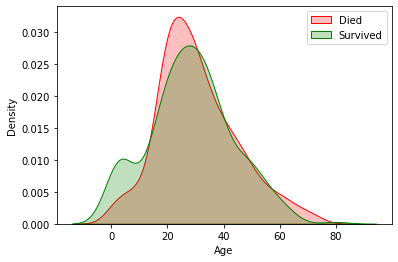
Did the age have a correlation with the chance of survival ?
plot = sns.kdeplot(df.loc[ df.Survived == 0, "Age"], color="Red", shade=True)
plot = sns.kdeplot(df.loc[ df.Survived == 1, "Age"], color="Green", shade=True)
plot.legend(["Died", "Survived"])
<matplotlib.legend.Legend at 0x178ccfa90>
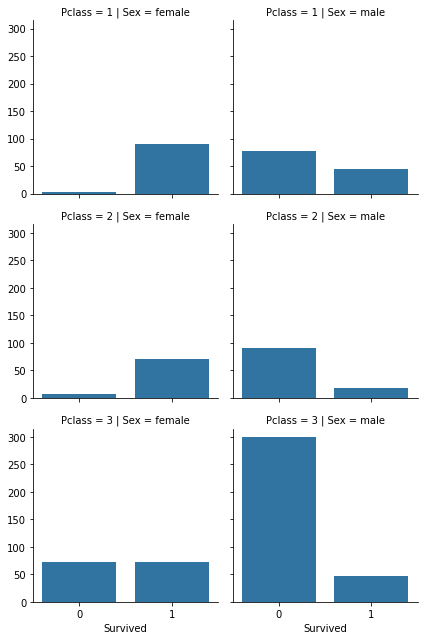
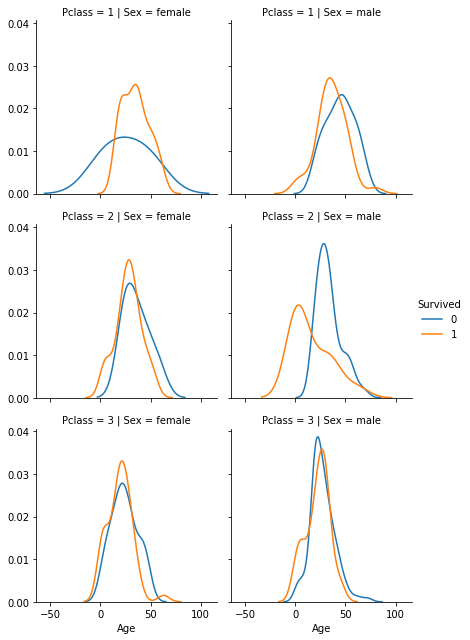
2 other plots (try to reproduce them):
- number of people for survivor and deceased person w.r.t. their Pclass and sex category
- distribution of ages for survivor and deceased person w.r.t their Pclass and sex
Missing values ?
df.isna().sum()
PassengerId 0
Survived 0
Pclass 0
Name 0
Sex 0
Age 177
SibSp 0
Parch 0
Fare 0
Cabin 687
Embarked 2
titleName 0
dtype: int64
Graphically (can be nice to see if some missing values in a column have corresponding missing values in other columns).
sns.heatmap(df.isna())
<AxesSubplot:>
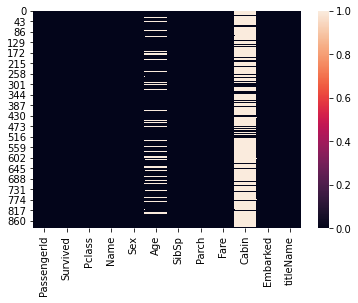
1. Handling missing values in Cabin column
df.Cabin.isna().sum()
687
df.Cabin.nunique()
147
a lot of unique different labels for Cabin + a lot of missing values for Cabin.
(687 + 147) / df.shape[0] * 100 # in %
93.60269360269359
df.drop("Cabin", axis=1, inplace=True)
2. Handling missing values in Embarked column
df.Embarked.isna().sum()
2
df.Embarked.value_counts()
S 644
C 168
Q 77
Name: Embarked, dtype: int64
df.Embarked.value_counts()[df.Embarked.mode()] / df.Embarked.value_counts().sum() *100
S 72.440945
Name: Embarked, dtype: float64
S level account for 72% of the values in the dataset.
Let’s replace the missing value by the mode (they are only 2 missing values in df.Embarked)
df.Embarked.fillna(df.Embarked.mode()[0], inplace=True)
df.Embarked.value_counts()
S 646
C 168
Q 77
Name: Embarked, dtype: int64
3. Handling missing values in Age column
sns.displot(df.Age, kde=True)
<seaborn.axisgrid.FacetGrid at 0x178cb5040>
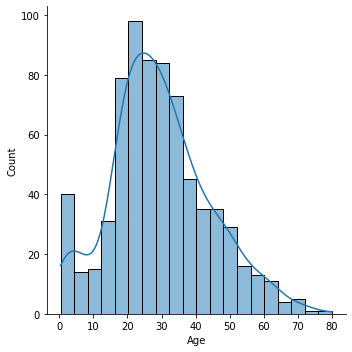
Replacing the age by the mean in the entire population is really a strong assumption, but i don’t want to put too much emphasis on this part.
df.Age.fillna(df.Age.mean(), inplace=True)
Modelling
final processing before injecting roughly in the model
df.drop("PassengerId", axis=1, inplace=True)
df.drop("Name", axis=1, inplace=True)
df.drop('flag', axis=1, inplace=True)
separating the target from the feature matrix:
X, y = df.drop("Survived", axis=1), df.Survived
encoding the categorical features
Some ML algorithm can’t accept non-encoded features as such.
- the Pclass is an ordinal variable (1st class > 2nd class > 3rd class)
from sklearn.preprocessing import OrdinalEncoder
encoder = OrdinalEncoder()
X[["Pclass"]] = encoder.fit_transform(X[["Pclass"]])
encoder.categories_
[array([1, 2, 3])]
- the sex column is not.
because there is no apparent ordering between male and female (e.g. can we say male > female or female > male ?)
same for Embarked and titleName (although we could argue about the later)
from sklearn.preprocessing import OneHotEncoder
encoder = OneHotEncoder(drop="first", sparse=False)
X[encoder.get_feature_names(["is", "embarked_from", "hastitle"])] =\
encoder.fit_transform(X[["Sex" , "Embarked", "titleName"]])
X.drop(["titleName", "Embarked", "Sex"], axis=1, inplace=True)
X.head()
| Pclass | Age | SibSp | Parch | Fare | is_male | embarked_from_Q | embarked_from_S | hastitle_Miss | hastitle_Mr | hastitle_Mrs | hastitle_other | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2.0 | 22.0 | 1 | 0 | 7.2500 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 |
| 1 | 0.0 | 38.0 | 1 | 0 | 71.2833 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 |
| 2 | 2.0 | 26.0 | 0 | 0 | 7.9250 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 |
| 3 | 0.0 | 35.0 | 1 | 0 | 53.1000 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 |
| 4 | 2.0 | 35.0 | 0 | 0 | 8.0500 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 |
Applying some models
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
from sklearn.linear_model import LogisticRegression
logreg = LogisticRegression(max_iter=1000)
logreg.fit(X_train, y_train)
LogisticRegression(max_iter=1000)
Let’s have a look at one metric: the accuracy
from sklearn.metrics import accuracy_score
accuracy_score(y_true=y_test, y_pred=logreg.predict(X_test))
0.776536312849162
doesn’t seem that bad, but… think again about the dataset itself and the proportion of each values y take.
from sklearn.metrics import plot_confusion_matrix
plot_confusion_matrix(logreg, X=X_test, y_true=y_test) # accept an already fitted model
<sklearn.metrics._plot.confusion_matrix.ConfusionMatrixDisplay at 0x1793eb490>
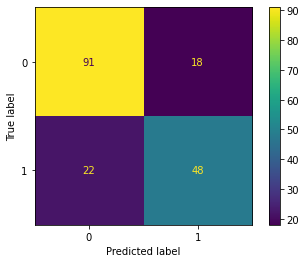
Accuracy is defined as the sum of the diagonal elements over the sum of all the elements of the confusion matrix.
In other words, which proportion of the observed y values match with the predictions made by the model, no matter what class (positive / negative) y belong too.
accuracy = (91+48)/(22+18+91+48)
accuracy
0.776536312849162
if we took a very extreme example where our model is “dummy” and only output y as died: then accuracy would be:
y_test.value_counts()[0] / y_test.value_counts().sum() *100
60.893854748603346
61%
Now, imagine we had 99% of people dying in the test set, the accuracy of this dummy model would raise up to 99 !.
Hence you should try to check some other metrics depending on the use-case, and/or the business mater, and/or whether you are in case of imbalanced dataset.
It often boils down to a trade-off:
- do you want to detect any true case of survival (
y_pred=y_obs=1) ? at the expense of predicting people as survivor (y_pred= 1) when they were actually observed as dead (y_obs=0). This is also named a false positive.- This case scenario would be a good use-case if we wanted to detect if someone could possibly have a rare disease. We would prefer the test to be overly detecting a disease even when the patient isn’t affected by any disease (
y_obs=0andy_pred=1): the patient could procede further tests to ensure it does not have the disease.
- This case scenario would be a good use-case if we wanted to detect if someone could possibly have a rare disease. We would prefer the test to be overly detecting a disease even when the patient isn’t affected by any disease (
- or do you prefer to detect any true case of death (
y_pred=y_obs=0)? at the expense of classifying people as dead (y_pred=0) when they were actually observed as survivor (y_obs=1). This is also named a false negative.- This case scenario would be a good use-case for spams detection. We certainly would want to detect spams, but one person may find it very annoying the emails he sends to people are flagged as spam by the AI engine (
y_pred=1) when it is clearly not (y_obs=0). So we even more want to prevent this from happening.
- This case scenario would be a good use-case for spams detection. We certainly would want to detect spams, but one person may find it very annoying the emails he sends to people are flagged as spam by the AI engine (
Of course in theory you could have a perfect model which does not create neither false positive, nor false negative, and output an accuracy of 100%. But this is not so often in practice.
Note that some metrics represent either of those scenarii, or combine (with some weighted proportion) a mix of both. I leave this to you as an exercice to find which is best suited to your problem..
Final words
- For the rest of this exercice you can try other models, once you have defined which metrics you want to assess your model performance.
- You can reuse the model validation techniques we used in other lessons (k-fold, gridsearch, learning curves, etc.)

 Coding a Neuron with Numpy
Coding a Neuron with Numpy