Why Pandas ?
To create a machine learning model, we need to apply the underlying algorithm on some training data (more on this in Lecture 5). For this to work, we need to have a specific data structure to pass as input.
Most traditional ML models require a 2D data-structure, just like a matrix. A numpy.array can be used for that purpose. Each row define an observation (more on this in Lecture 5), types of observations might depend on our designed problem. Each column display a caracteristic for each of these observations.
Now, imagine such 2D data structure, maybe you would want to first name your columns and rows, analogously to a spreadsheet or SQL table, then inspect your data, handle missing values, do some processing on it (e.g. retrieve number of streets out of the street name), combine with other related, 2D, data from different sources, quickly perform descriptive statistics, do some computations on columns, on rows, or even within groups of observations sharing some common arbitrary caracteristic, quickly display trends from your computations.
Also, dealing with initially less structured, clean and complete data, consists in most of the time spent by the data scientist. Sometimes the data you’re being given doesn’t have such 2D representation and you would want to have some helpers functions to perform the conversion.
In either cases, Pandas package and its DataFrame object comes in handy.
Three important Pandas’ data-structures
From the pandas docs, which give a nice overview of the package:
pandas.DataFrameis a 2-dimensional labeled data structure with columns of potentially different types. You can think of it like a spreadsheet. It got rows and columns’ labels (pandas.Indexobjects). Each column is apandas.Series.
Pandas is built on top of Numpy, hence sharing some optimizations from the latter, as well as closely related API.
In the rest of this tutorial we will mainly work on the DataFrame class, although we first need to introduce the 2 other core data structures mentioned sooner: the Series and the Index, as they are each constitutive of DataFrame and the former share also similarly named methods and behaviors with the DataFrame class.
Series
Definition
one-dimensional array of indexed data.
pd.Series([3,2,1])
0 3
1 2
2 1
dtype: int64
with explicit index definition !
Example:
serie_1 = pd.Series([3,2,1], index=[93,129, 219394])
serie_1
93 3
129 2
219394 1
dtype: int64
serie_1.index
Int64Index([93, 129, 219394], dtype='int64')
Series as a dictionnary-like object
A dictionnary-like, object with possible keys index repetition.
serie = pd.Series([3,2,1], index=["rené", "rené", "jean"])
We display a series.
serie
rené 3
rené 2
jean 1
dtype: int64
We can show the values, a numpy typed array.
serie.values
array([3, 2, 1])
and show the keys index !
serie.index
Index(['rené', 'rené', 'jean'], dtype='object')
Here the 4 basic dict operations work seamlessly the same for a Series.
- Access by
keyindex:
serie['rené']
rené 3
rené 2
dtype: int64
- Set a new
keyindex:value pair
serie['joseph'] = 5
- Change a value for a given
keyindex.
serie['rené'] = 4
Notice the broadcoasting of the integer here in case of multiple same index for the given value.
serie
rené 4
rené 4
jean 1
joseph 5
dtype: int64
You can also pass a sequence of elements with a matching length with the index multiplicity number.
serie['rené'] = [4,3]
serie
rené 4
rené 3
jean 1
joseph 5
dtype: int64
- delete a
keyindex:value pair
del serie["rené"]
serie
jean 1
joseph 5
dtype: int64
You can also do lookups on indexes, using the same syntax as for dict keys.
print('rené' in serie) #in the indexes, same syntax as for dict keys
print("jean" in serie)
False
True
- When index is unique, pandas use a hashtable just like
dicts : O(1). - When index is non-unique and sorted, pandas use binary search O(logN)
- When index is non-unique and not-sorted, pandas need to check all the keys just like a list look-up: O(N).
You can also do some other things you would not be able using dict primitive, like slicing.
serie[0:4:2] # indexing: not possible in a simple dict
jean 1
dtype: int64
The similarity with dict is although so close you can use a dict in the pd.Series constructor. This automatically create the indexes from the keys in the dict and the values from the corresponding values in the dict.
Note: the index:value order in the newly created pd.Series can be slightly different for different concomittent versions of Python and Pandas. Pandas >= 0.23 conserve the insertion order from the underlying dict argument, although you still Python versions above 3.6 to maintain dict keys’insertion order (use OrderedDict for versions before).
test = pd.Series(dict(zip(["ea","fzf","aeif"], [2,3,2])))
# with zip or using a dict
test2 = pd.Series({"ea":2, "fzf":3, "aeif":2}, index=["ea"])
test
aeif 2
ea 2
fzf 3
dtype: int64
test2
ea 2
dtype: int64
If multiple different types reside in a Series, all of the data will get upcasted to a dtype that accommodates all of the data involved.
test2 = pd.Series({"ea":2, "fzf":3, "aeif":"zf"}, index=["ea"])
test2
ea 2
dtype: object
test2 = pd.Series({"ea":2, "fzf":3, "aeif":2.4}, index=["ea"])
test2
ea 2.0
dtype: float64
dtype=object means that the best common type infered representation for the contents of the pd.Series is “a Python object”. (Everything is object in Python see Lecture 2!).
This also means a performance drop, any operations on the data will be done at the Python level. Python for-loops will be performed, checking the actual type of each ‘object’ for the operation one want to perform on the input vector (1)
Selection in Series
Masking
A Series mask is a Series as a collection of indexes:boolean-values, which can be later used to filter-out elements from another Series, based on the falsy evaluated values for each index in the former.
Performing a comparison on a Series creates a mask of same shape, with indexes from the original array along with true or false results originating from the element-wise comparisons from your original comparison expression.
(test>2)
aeif False
ea False
fzf True
dtype: bool
(test<4)
aeif True
ea True
fzf True
dtype: bool
Since numpy arrays support vectorized calculations (more on that later) and does not contain arbitrary unlike typed elements as for lists, you can use the & bitwise operator, a element-wise version of the logical and.
# not "and" but "&" : & operator is a bitwise "and"
(test>2) & (test < 4)
aeif False
ea False
fzf True
dtype: bool
You can see the result is still a Series, but this time of boolean values (check the dtype !)
type((test>2) & (test < 4) )
pandas.core.series.Series
We can later keep it in as a ‘mask’ variable, it is particularly useful when a lot comparisons should be given a meaningful name (e.g. mask variable here could be lower4greater2 for example).
# mask ( the last expression whose result is an pd.Serie stored in the variable mask)
mask = (test>2) & (test < 4)
test[mask]
fzf 3
dtype: int64
Indexing
We can also select a value for a given index, as highlighted in the introductory section on Series.
Although we should take extra care when doing so: the previous notation, e.g. serie['rené'], makes use of the names of the indexes we explicitly defined earlier.
Fancy indexing
This is just a fancy word for selecting multiple indexes, provisioning a list of indexes.
# fancy indexing (<=> selecting multiple indexes using a list of indexes)
test[["ea", "fzf"]]
ea 2
fzf 3
dtype: int64
Slicing and the confusion of explicit vs implicit indexes
This is another word for selecting a subset of an original array (or even list), based on an interval constructed using a start, stop and [step] elements.
For Series, we can slice a Series by 2 ways:
- using the names we explicitly defined (or defaulted as integer-based indexes creation) for the index at
Seriescreation time, for start and stop values. -
using implicit start and stop values for the index. By implicit we mean integers which define the order of appearance of the index itself, taking caution that the first element is of index 0 in Python.
- Explicit index slicing:
# (using the labels of the indexes)
test["aeif": "fzf"]
aeif 2
ea 2
fzf 3
dtype: int64
- Implicit index slicing (using integers i.e. order of appearance):
test[0: 2]
aeif 2
ea 2
dtype: int64
-
using explicit indexes while slicing include the final index
-
using implicit index in slicing exclude the final index
What about i defined at creation time a Series with integer index values and i want to slice them ? 🙄
serie2 = pd.Series({1:4, 2:8, 3:51})
serie2
1 4
2 8
3 51
dtype: int64
Here you see that for indexing, the explicit index is used, i.e. “element of index defined as 3”, but for slicing it is takes elements from the 2nd indexed element to the 3rd, excluded, indexed element, no matter what value of index the elements are.
serie2[3] # indexing: defaults to select explicit index / with label 3
serie2[2:3] # slicing: defaults to select implicit indexes
51
3 51
dtype: int64
Loc and Iloc accessors
These accessors give a great alternative from default, albeit confusing, slicing behaviors with respect to the indexes’values.
Using loc property:
This forces indexing and slicing using the explicitly defined index values:
serie2.loc[1] # indexing: on explicit index
serie2.loc[2:3] # slicing: on explicit index
4
2 8
3 51
dtype: int64
Using iloc property:
This forces indexing and slicing using implicit indexes i.e. order of appearance of the elements from 0 (1st element) to n-1 (last one). This also means you will never use something else in iloc accessors than integers.
serie2.iloc[1] # indexing: on implicit index
serie2.iloc[2:3] # slicing: on implicit index
8
3 51
dtype: int64
serie2.loc[1:5] # slicing: explicit index
serie2.iloc[1:5] # slicing: implicit index
1 4
2 8
3 51
dtype: int64
2 8
3 51
dtype: int64
serie2.loc[[1,2]] # fancy indexing
serie2.iloc[[1,2]] # fancy indexing
1 4
2 8
dtype: int64
2 8
3 51
dtype: int64
dtypes
A serie being an-indexed array, it then maintain a well-known implementation feature from numpythat infers about the overall representation of the data within the array: the dtype.
Recall that infering the data representation which accomodates all elements in the array is what enables, in case, for example, of numerical data (integers, float), to not only stored the values as C integers (avoiding the overhead introduced by Python primitive types) but also to perform efficient operations on the C for-loop level, as it is not required to dynamically type-check every single elements of the array and find which function to dispatch accordingly.
%timeit np.arange(1E6, dtype="int").sum()
1.14 ms ± 142 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)
%timeit np.arange(1E6, dtype="float").sum()
1.25 ms ± 17.7 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)
s = pd.Series(['The', 3, 'brown', 'fox'])
s*2
0 TheThe
1 6
2 brownbrown
3 foxfox
dtype: object
s.values
array(['The', 3, 'brown', 'fox'], dtype=object)
Link. Creating an array with dtype=object is different. The memory taken by the array now is filled with pointers to Python objects which are being stored elsewhere in memory (much like a Python list is really just a list of pointers to objects, not the objects themselves).
%timeit np.arange(1E6, dtype="object").sum()
77.8 ms ± 4.15 ms per loop (mean ± std. dev. of 7 runs, 10 loops each)
Index object
can be sliced or indexed
…just like an array, because it is indeed an one-dimensional array.
serie2.index[0]
1
serie2.index[:2]
Int64Index([1, 2], dtype='int64')
have sets’ operations
By this we mean it does have common bitwise operatoin
serie2.index & {1, 5}
Int64Index([1], dtype='int64')
serie2.index ^ {1,5}
Int64Index([2, 3, 5], dtype='int64')
are immutables
serie2.index[0]=18
Traceback (most recent call last):
File "<ipython-input-1096-707f9cda8675>", line 1, in <module>
serie2.index[0]=18
File "/Users/lucbertin/.pyenv/versions/3.5.7/lib/python3.5/site-packages/pandas/core/indexes/base.py", line 4260, in __setitem__
raise TypeError("Index does not support mutable operations")
TypeError: Index does not support mutable operations
DataFrame
The “main thing” of this lecture that we are going to use intensively in the rest of this lecture:
-
sequence of “aligned” Series objects (sharing the same indexes / like an Excel file).
-
each Series object is a column.
-
Hence
pd.DataFramecan be seen as dictionnary of Series objects. -
Flexible rows and columns’ labels (
Indexobjects for both).
Construction
serie1 = pd.Series({"Luc": 25, "Corentin":29, "René": 40})
serie2 = pd.Series({"René": "100%", "Corentin": "25%", "Luc": "20%"})
# dictionnary of pd.Series
df = pd.DataFrame({"note": serie1,
"charge_de_travail": serie2})
df
| charge_de_travail | note | |
|---|---|---|
| Corentin | 25% | 29 |
| Luc | 20% | 25 |
| René | 100% | 40 |
# index objects on both columns and rows
df.index
df.columns
Index(['Corentin', 'Luc', 'René'], dtype='object')
Index(['charge_de_travail', 'note'], dtype='object')
If you pass an index and / or columns, you are guaranteeing the index and / or columns of the resulting DataFrame. Thus, a dict of Series plus a specific index will discard all data not matching up to the passed index.
df2 = pd.DataFrame({"note": serie1,
"charge_de_travail": serie2},
index=["Corentin", "Luc", "Julie"],
columns=["note", "autre"])
df2
# filled with NaN ("Not A Number")
# when no value exist for the given (row_index, column_index)
| note | autre | |
|---|---|---|
| Corentin | 29.0 | NaN |
| Luc | 25.0 | NaN |
| Julie | NaN | NaN |
The DataFrame can be constructed using a list of dictionary.
- each
dictelement is a row. - each key of each dict refers to a column.
df2 = pd.DataFrame([{'a': 1, 'b': 2}, {'b': 3, 'c': 4}])
df2
| a | b | c | |
|---|---|---|---|
| 0 | 1.0 | 2 | NaN |
| 1 | NaN | 3 | 4.0 |
pd.DataFrame([(1, 1, 3), (1, 2,4), (1,1,1)],
columns=["a", "b", "c"],
index=["Jean", "Jacques", "René"])
| a | b | c | |
|---|---|---|---|
| Jean | 1 | 1 | 3 |
| Jacques | 1 | 2 | 4 |
| René | 1 | 1 | 1 |
You can also create a DataFrame by using pandas methods for reading supported file format, e.g. using pd.read_csv method.
Shape property
df.shape
(3, 2)
shape: tuple of the number of elements with respect to each dimension
-
For a 1D array, the shape would be (n,) where n is the number of elements in your array.
-
For a 2D array, the shape would be (n,m) where n is the number of rows and m is the number of columns in your array
accessing a column/Serie by key :
Accessing columns
As DataFrame is seen like a dictionary of Series / columns, you can access one of them using the corresponding key column’index !
df['note']
Corentin 29
Luc 25
René 40
Name: note, dtype: int64
Using the attribute notation is not advised especially for assignements as some methods or attributes of the same name already exist in the DataFrame class’ own namespace.
df.note
Corentin 29
Luc 25
René 40
Name: note, dtype: int64
Indexing or Slicing
Indexing works the same way as for Series, but you have to account this time for the second dimension
df.loc_or_iloc[ dim1 = rows, dim2 = columns]
df.iloc[:3, :1] # implicit indexing
| charge_de_travail | |
|---|---|
| Corentin | 25% |
| Luc | 20% |
| René | 100% |
columns slicing/indexing is optional here, without specifying it, you select only rows
df.iloc[:3]
| charge_de_travail | note | |
|---|---|---|
| Corentin | 25% | 29 |
| Luc | 20% | 25 |
| René | 100% | 40 |
df.loc["Corentin":"Luc","charge_de_travail":"note"] # explicit indexing
| charge_de_travail | note | |
|---|---|---|
| Corentin | 25% | 29 |
| Luc | 20% | 25 |
same thing here, only rows selected
df.loc[:"Corentin"]
| charge_de_travail | note | |
|---|---|---|
| Corentin | 25% | 29 |
df.loc[["Corentin", "Luc"], :] # mixing slicing and fancy indexing
| charge_de_travail | note | |
|---|---|---|
| Corentin | 25% | 29 |
| Luc | 20% | 25 |
Something to mention here: by default, without using accessors like loc and iloc, indexing or fancy indexing directly df, performs the indexing on its columns.
df[["charge_de_travail"]] # indexing directly df defaults to columns
| charge_de_travail | |
|---|---|
| Corentin | 25% |
| Luc | 20% |
| René | 100% |
Finally, of course you can set a new value for an element on some (row_index, column_index) using either of those accessors:
df.iloc[0,2] = np.nan
Masking
You can also use masking here and draw comparisons on a Dataframe level (e.g. df > 3), or on Series/column level, e.g. df["sexe"] == "Homme", df["age"] > 18.
In the first case, the resulting object will be a DataFrame filled with boolean values. In the second one, as before, a Series with boolean values.
A difference here using Series as a mask is that filtering a DataFrame using only true evaluated values from a Series (a 1D indexed-array then), keeps the entire rows as you may have multiple aligned Series/ columns for one given Index (with a true value).
Slicing using slice notation (::), or masking is performed on rows by default.
mask = df["charge_de_travail"]=="25%"
mask
Corentin True
Luc False
René False
Name: charge_de_travail, dtype: bool
Note that the Series-like mask, having the explictly defined indexes from the original one, you can still use df.loc upon filtering.
df[mask] # masking directly df is operated on rows
# same as df.loc[mask]
| charge_de_travail | note | |
|---|---|---|
| Corentin | 25% | 29 |
df[:3] # slicing directly df is operated on rows
| charge_de_travail | note | |
|---|---|---|
| Corentin | 25% | 29 |
| Luc | 20% | 25 |
| René | 100% | 40 |
Operations between DataFrames
What about multiplying all elements from a DataFrame by 2?
What about adding 2 DataFrame ?
We first need to distinct 2 types of operations into binary and unary operations:
- 3 - 2 <=> substract(3,2) <=> binary operation (2 inputs)
- -2 <=> neg(2) <=> unary operation (one input)
- sin(2) <=> unary operation (one input)
in Pandas :
- unary operations on
dfs elements preserve the indexes. - binary operations on elements from 2
dfs align the operations on the indexes. These behaviors come from numpyufuncs(universal functions i.e. vectorized functions i.e. that take the whole vector as input, applying the function element-wise) which can be used forDataFramess too.
import numpy as np
rng = np.random.RandomState(42) # for reproducibility
data = rng.randint(0,10, (3,4)) # creating an array of random integer values
df = pd.DataFrame(data)
df
| 0 | 1 | 2 | 3 | |
|---|---|---|---|---|
| 0 | 6 | 3 | 7 | 4 |
| 1 | 6 | 9 | 2 | 6 |
| 2 | 7 | 4 | 3 | 7 |
df2 = pd.DataFrame(rng.randint(0,10, (4,4)))
df2
| 0 | 1 | 2 | 3 | |
|---|---|---|---|---|
| 0 | 7 | 2 | 5 | 4 |
| 1 | 1 | 7 | 5 | 1 |
| 2 | 4 | 0 | 9 | 5 |
| 3 | 8 | 0 | 9 | 2 |
We will use for the later example of summation between 2 DataFrames, reindex just to rearranged indexes of df2 (this does not change the association indexed-value !)
df2 = df2.reindex([1,0,2,3]) #just to show rearranged indexes (does not change the association with the indexed data)
df2
| 0 | 1 | 2 | 3 | |
|---|---|---|---|---|
| 1 | 1 | 7 | 5 | 1 |
| 0 | 7 | 2 | 5 | 4 |
| 2 | 4 | 0 | 9 | 5 |
| 3 | 8 | 0 | 9 | 2 |
df + df2
| 0 | 1 | 2 | 3 | |
|---|---|---|---|---|
| 0 | 13.0 | 5.0 | 12.0 | 8.0 |
| 1 | 7.0 | 16.0 | 7.0 | 7.0 |
| 2 | 11.0 | 4.0 | 12.0 | 12.0 |
| 3 | NaN | NaN | NaN | NaN |
on line of index 0, 7+6 = 13
which shows indexes had been aligned during the binary operation
also notice the union of the indices during the binary operation. If one may not exist in either of the dataframes and the result can’t be evalutated, NaN fill the concerned entries
df.__add__(df2, fill_value=25) # used in the binary operation 25+8 = 33)
| 0 | 1 | 2 | 3 | |
|---|---|---|---|---|
| 0 | 13.0 | 5.0 | 12.0 | 8.0 |
| 1 | 7.0 | 16.0 | 7.0 | 7.0 |
| 2 | 11.0 | 4.0 | 12.0 | 12.0 |
| 3 | 33.0 | 25.0 | 34.0 | 27.0 |
Operations between Series and Dataframe
Performing an operation between a Serie and a DataFrame implies performing an operation between data structures of different shapes, hence implying numpy broadcasting.
From the Numpy docs:
Broadcasting is how numpy treats arrays with different shapes during arithmetic operations. Subject to certain constraints, the smaller array is “broadcast” across the larger array so that they have compatible shapes. Broadcasting provides a means of vectorizing array operations so that looping occurs in C instead of Python
The only requirement for broadcasting is a way aligning array dimensions such that either :
- aligned dimensions are equal (so that operations are done on an element-by-element basis from 2 arrays of same shape)
- one of the aligned dimensions is 1 (in other words, dimensions with size 1 are stretched or “copied” to match the dimension of the other array)
Operations between pandas.Series and pandas.DataFram then respect the numpy broadcasting rules:
If the two arrays differ in their number of dimensions, the shape of the one with fewer dimensions is padded with ones on its leading (left) side.’ (2)
df.shape, df.iloc[1].shape, df.iloc[1][np.newaxis, :].shape
((3, 4), (4,), (1, 4))
df
df.iloc[1]
| 0 | 1 | 2 | 3 | |
|---|---|---|---|---|
| 0 | 6 | 3 | 7 | 4 |
| 1 | 6 | 9 | 2 | 6 |
| 2 | 7 | 4 | 3 | 7 |
0 6
1 9
2 2
3 6
Name: 1, dtype: int64
df - df.iloc[1] #row-wise (1,4) copied other 3 times => (3,4)
| 0 | 1 | 2 | 3 | |
|---|---|---|---|---|
| 0 | 0 | -6 | 5 | -2 |
| 1 | 0 | 0 | 0 | 0 |
| 2 | 1 | -5 | 1 | 1 |
df - df.iloc[1].sample(4) # again: kept the index alignements during computation
| 0 | 1 | 2 | 3 | |
|---|---|---|---|---|
| 0 | 0 | -6 | 5 | -2 |
| 1 | 0 | 0 | 0 | 0 |
| 2 | 1 | -5 | 1 | 1 |
if you want to do it columnwise and not row wise
df.__sub__(df.iloc[1], axis=0) # caution, the indexes operations will be based on the column indexes
| 0 | 1 | 2 | 3 | |
|---|---|---|---|---|
| 0 | 0.0 | -3.0 | 1.0 | -2.0 |
| 1 | -3.0 | 0.0 | -7.0 | -3.0 |
| 2 | 5.0 | 2.0 | 1.0 | 5.0 |
| 3 | NaN | NaN | NaN | NaN |
df.columns = ["a","b",0,"d"]
df
| a | b | 0 | d | |
|---|---|---|---|---|
| 0 | 6 | 3 | 7 | 4 |
| 1 | 6 | 9 | 2 | 6 |
| 2 | 7 | 4 | 3 | 7 |
df.iloc[1]
a 6
b 9
0 2
d 6
Name: 1, dtype: int64
df.__sub__(df.iloc[1], axis=0)
# based on the column indexes
# only 0 match with one of the column index label
| a | b | 0 | d | |
|---|---|---|---|---|
| 0 | 4.0 | 1.0 | 5.0 | 2.0 |
| 1 | NaN | NaN | NaN | NaN |
| 2 | NaN | NaN | NaN | NaN |
| a | NaN | NaN | NaN | NaN |
| b | NaN | NaN | NaN | NaN |
| d | NaN | NaN | NaN | NaN |
Close API with Series and numpy
The DataFrame object, constituted of Series object and being also an ‘enhanced version’ of a numpy array, no wonder why a major part of the API for one can be reused for the other.
Here are just example of reused methods or properties (you’ve seen other ones in the course like loc and iloc for instance):
df[0].shape, df.shape
((3,), (3, 4))
df2 = df - pd.DataFrame([(1,2), (4,5), (9,19)], columns=["a","b"])
df2
| 0 | a | b | d | |
|---|---|---|---|---|
| 0 | NaN | 5 | 1 | NaN |
| 1 | NaN | 2 | 4 | NaN |
| 2 | NaN | -2 | -15 | NaN |
print(df.dtypes)
print(df2.dtypes)
# NaN is a floating-point value,
# hence the Series embedding it gets its dtype upcasted to float (if it were an int)
# this pd.Series supports fast operations contrarily to a Series of dtype=object
# because Python needs to type check dynamically every time
a int64
b int64
0 int64
d int64
notes object
dtype: object
Name object
id_account object
id_client object
dtype: object
Managing missing values
This is a real asset of pandas Dataframe in comparison to numpy(which would find other strategies for handling missing values during computations, using classes like “masked arrays”).
Managing missing values, either by dropping or imputing them based on some type of criteria, is an crucial step you should always document during your data scientist experiments. It is always a hot spot in areas such as Statistics or Machine Learning.
A mishandling of NA values can definitely have an impact on your results or greatly lower your model performance trained on the data.
pd.Series([2, np.nan]).isnull()
0 False
1 True
dtype: bool
df2.iloc[0,2] = np.nan
df2
df2.isnull()
| 0 | a | b | d | |
|---|---|---|---|---|
| 0 | NaN | 5 | NaN | NaN |
| 1 | NaN | 2 | 4.0 | NaN |
| 2 | NaN | -2 | -15.0 | NaN |
| 0 | a | b | d | |
|---|---|---|---|---|
| 0 | True | False | True | True |
| 1 | True | False | False | True |
| 2 | True | False | False | True |
pd.Series([2, np.nan]).dropna()
0 2.0
dtype: float64
df2
df2.dropna(axis=1) # drop a column when contains one NA value
df2.dropna(axis=0) # drop a row when contains one NA value
df2.dropna(axis=1, how="all") # drop a column when contains all NA value
df2.dropna(axis=1, thresh=3) # drop a column if below 3 non-NA value
| 0 | a | b | d | |
|---|---|---|---|---|
| 0 | NaN | 5 | NaN | NaN |
| 1 | NaN | 2 | 4.0 | NaN |
| 2 | NaN | -2 | -15.0 | NaN |
| a | |
|---|---|
| 0 | 5 |
| 1 | 2 |
| 2 | -2 |
| 0 | a | b | d |
|---|
| a | b | |
|---|---|---|
| 0 | 5 | NaN |
| 1 | 2 | 4.0 |
| 2 | -2 | -15.0 |
| a | |
|---|---|
| 0 | 5 |
| 1 | 2 |
| 2 | -2 |
df2
df2.fillna(value=2) #fill NA with specified value
# fill NA backwards
# i.e. using the following non-null element
# to fill preceding NA ones
# defaults on rows basis
df2.fillna(method="bfill")
df2.fillna(method="bfill", axis=1) # on column basis
| 0 | a | b | d | |
|---|---|---|---|---|
| 0 | NaN | 5 | NaN | NaN |
| 1 | NaN | 2 | 4.0 | NaN |
| 2 | NaN | -2 | -15.0 | NaN |
| 0 | a | b | d | |
|---|---|---|---|---|
| 0 | 2.0 | 5 | 2.0 | 2.0 |
| 1 | 2.0 | 2 | 4.0 | 2.0 |
| 2 | 2.0 | -2 | -15.0 | 2.0 |
| 0 | a | b | d | |
|---|---|---|---|---|
| 0 | NaN | 5 | 4.0 | NaN |
| 1 | NaN | 2 | 4.0 | NaN |
| 2 | NaN | -2 | -15.0 | NaN |
| 0 | a | b | d | |
|---|---|---|---|---|
| 0 | 5.0 | 5.0 | NaN | NaN |
| 1 | 2.0 | 2.0 | 4.0 | NaN |
| 2 | -2.0 | -2.0 | -15.0 | NaN |
MultiIndex
data = {('group1', 'Luc'): 18,
('group2', 'Jean'): 23,
('group1', 'Seb'): 17,
('group1', 'René'): 4,
('group2', 'Alex'): 4,
('group3', 'Sophie'): 25,
('group2', 'Camille'): 2 }
serie = pd.Series(data)
serie
group1 Luc 18
René 4
Seb 17
group2 Alex 4
Camille 2
Jean 23
group3 Sophie 25
dtype: int64
serie[:,"Luc"]
group1 18
dtype: int64
serie["group1"]
Luc 18
René 4
Seb 17
dtype: int64
serie[serie>=18]
group1 Luc 18
group2 Jean 23
group3 Sophie 25
dtype: int64
# creating the multi-index using cartesian product
index = pd.MultiIndex.from_arrays([['group1', 'a', 'b', 'b'], ["Luc", 2, 1, 2]])
serie.reindex(index) # works for multi-index too !
# Conform Series to new index with optional filling logic, placing
# NA/NaN in locations having no value in the previous index
group1 Luc 18.0
a 2 NaN
b 1 NaN
2 NaN
dtype: float64
# hierarchical indices and columns
index = pd.MultiIndex.from_product([[2013, 2014], [1, 2]],
names=['year', 'visit'])
columns = pd.MultiIndex.from_product(
[['Bob', 'Guido', 'Sue'], ['HR', 'Temp']],names=['subject', 'type'])
# mock some data
data = np.round(np.random.randn(4, 6), 1)
data[:, ::2] *= 10
data += 37
# create the DataFrame
health_data = pd.DataFrame(data, index=index, columns=columns)
health_data
| subject | Bob | Guido | Sue | ||||
|---|---|---|---|---|---|---|---|
| type | HR | Temp | HR | Temp | HR | Temp | |
| year | visit | ||||||
| 2013 | 1 | 52.0 | 36.4 | 38.0 | 36.6 | 32.0 | 38.1 |
| 2 | 28.0 | 37.7 | 47.0 | 35.4 | 50.0 | 36.4 | |
| 2014 | 1 | 30.0 | 37.0 | 16.0 | 36.6 | 49.0 | 37.7 |
| 2 | 52.0 | 36.8 | 31.0 | 35.5 | 36.0 | 37.6 | |
health_data.loc[:2013 , ("Bob")]
| type | HR | Temp | |
|---|---|---|---|
| year | visit | ||
| 2013 | 1 | 52.0 | 36.4 |
| 2 | 28.0 | 37.7 |
#health_data.loc[(:,1),["Bob"]] # can't use the tuple to define index
idx = pd.IndexSlice
health_data.loc[idx[:, 1], idx[:, 'HR']]
| subject | Bob | Guido | Sue | |
|---|---|---|---|---|
| type | HR | HR | HR | |
| year | visit | |||
| 2013 | 1 | 52.0 | 38.0 | 32.0 |
| 2014 | 1 | 30.0 | 16.0 | 49.0 |
Unstacking and Stacking: a matter of dimensionality
Creating a multiIndex rather than a simple Index is like creating an extra-dimension in our dataset.
We can take for each year, a 2D sub-dataframe composed of Bob’s HR visits.
This DataFrame hence can be seen as having 4 dimensions.
we can go back and forth from a multi-index series to a dataframe using unstack, so that one of the index level occupies the extra dimension given by the transition to a DataFrame
serie
group1 Luc 18
René 4
Seb 17
group2 Alex 4
Camille 2
Jean 23
group3 Sophie 25
dtype: int64
serie.unstack() #level -1 by default = most inner one
| Alex | Camille | Jean | Luc | René | Seb | Sophie | |
|---|---|---|---|---|---|---|---|
| group1 | NaN | NaN | NaN | 18.0 | 4.0 | 17.0 | NaN |
| group2 | 4.0 | 2.0 | 23.0 | NaN | NaN | NaN | NaN |
| group3 | NaN | NaN | NaN | NaN | NaN | NaN | 25.0 |
df3 = serie.unstack(level=0)
df3
| group1 | group2 | group3 | |
|---|---|---|---|
| Alex | NaN | 4.0 | NaN |
| Camille | NaN | 2.0 | NaN |
| Jean | NaN | 23.0 | NaN |
| Luc | 18.0 | NaN | NaN |
| René | 4.0 | NaN | NaN |
| Seb | 17.0 | NaN | NaN |
| Sophie | NaN | NaN | 25.0 |
# to reset the index and create it as a simple new column you can use reset_index()
df3.reset_index()
| index | group1 | group2 | group3 | |
|---|---|---|---|---|
| 0 | Alex | NaN | 4.0 | NaN |
| 1 | Camille | NaN | 2.0 | NaN |
| 2 | Jean | NaN | 23.0 | NaN |
| 3 | Luc | 18.0 | NaN | NaN |
| 4 | René | 4.0 | NaN | NaN |
| 5 | Seb | 17.0 | NaN | NaN |
| 6 | Sophie | NaN | NaN | 25.0 |
You can do some aggregation by index level (we are going to see this extensively on GroupBy section
health_data.mean(level='year')
health_data.mean(level='visit')
health_data.mean(axis=1, level='type')
| subject | Bob | Guido | Sue | |||
|---|---|---|---|---|---|---|
| type | HR | Temp | HR | Temp | HR | Temp |
| year | ||||||
| 2013 | 40.0 | 37.05 | 42.5 | 36.00 | 41.0 | 37.25 |
| 2014 | 41.0 | 36.90 | 23.5 | 36.05 | 42.5 | 37.65 |
| subject | Bob | Guido | Sue | |||
|---|---|---|---|---|---|---|
| type | HR | Temp | HR | Temp | HR | Temp |
| visit | ||||||
| 1 | 41.0 | 36.70 | 27.0 | 36.60 | 40.5 | 37.9 |
| 2 | 40.0 | 37.25 | 39.0 | 35.45 | 43.0 | 37.0 |
| type | HR | Temp | |
|---|---|---|---|
| year | visit | ||
| 2013 | 1 | 40.666667 | 37.033333 |
| 2 | 41.666667 | 36.500000 | |
| 2014 | 1 | 31.666667 | 37.100000 |
| 2 | 39.666667 | 36.633333 |
Concatenating DataFrames
pd.concat is here for the rescue !
df1
df2
| 0 | a | b | d | |
|---|---|---|---|---|
| 0 | 2.0 | False | 2.0 | 2.0 |
| 1 | 2.0 | False | 0.0 | 2.0 |
| 2 | 2.0 | False | 0.0 | 2.0 |
| 0 | a | b | d | |
|---|---|---|---|---|
| 0 | NaN | 5 | NaN | NaN |
| 1 | NaN | 2 | 4.0 | NaN |
| 2 | NaN | -2 | -15.0 | NaN |
pd.concat([df1, df2], axis=0) # concatenate rows (default)
pd.concat([df1, df2], axis=1) # concatenate columns (default)
| 0 | a | b | d | |
|---|---|---|---|---|
| 0 | 2.0 | 0 | 2.0 | 2.0 |
| 1 | 2.0 | 0 | 0.0 | 2.0 |
| 2 | 2.0 | 0 | 0.0 | 2.0 |
| 0 | NaN | 5 | NaN | NaN |
| 1 | NaN | 2 | 4.0 | NaN |
| 2 | NaN | -2 | -15.0 | NaN |
| 0 | a | b | d | 0 | a | b | d | |
|---|---|---|---|---|---|---|---|---|
| 0 | 2.0 | False | 2.0 | 2.0 | NaN | 5 | NaN | NaN |
| 1 | 2.0 | False | 0.0 | 2.0 | NaN | 2 | 4.0 | NaN |
| 2 | 2.0 | False | 0.0 | 2.0 | NaN | -2 | -15.0 | NaN |
the indices are preserved, even duplicated
verify_integrity=True can check if index from each df are differents
ignore_index=True just override the indexes after concatenation by a new integer one
keys = ["source1", "source2"] leave the indexes as is but create a new outer level from the 2 different sources/df of the data concatenated
pd.concat([df1, df2], axis=0, keys=["source1", "source2"])
| 0 | a | b | d | ||
|---|---|---|---|---|---|
| source1 | 0 | 2.0 | 0 | 2.0 | 2.0 |
| 1 | 2.0 | 0 | 0.0 | 2.0 | |
| 2 | 2.0 | 0 | 0.0 | 2.0 | |
| source2 | 0 | NaN | 5 | NaN | NaN |
| 1 | NaN | 2 | 4.0 | NaN | |
| 2 | NaN | -2 | -15.0 | NaN |
join='inner' keeps only the columns in common from the concatenation
df2["note"] = 2
df2
| 0 | a | b | d | note | |
|---|---|---|---|---|---|
| 0 | NaN | 5 | NaN | NaN | 2 |
| 1 | NaN | 2 | 4.0 | NaN | 2 |
| 2 | NaN | -2 | -15.0 | NaN | 2 |
pd.concat([df1, df2], axis=0, join='inner')
| 0 | a | b | d | |
|---|---|---|---|---|
| 0 | 2.0 | 0 | 2.0 | 2.0 |
| 1 | 2.0 | 0 | 0.0 | 2.0 |
| 2 | 2.0 | 0 | 0.0 | 2.0 |
| 0 | NaN | 5 | NaN | NaN |
| 1 | NaN | 2 | 4.0 | NaN |
| 2 | NaN | -2 | -15.0 | NaN |
serie1
serie2
serie1.append(serie2)
Corentin 29
Luc 25
René 40
dtype: int64
Corentin 25%
Luc 20%
René 100%
dtype: object
Corentin 29
Luc 25
René 40
Corentin 25%
Luc 20%
René 100%
dtype: object
df1.append(df2)
/Users/lucbertin/.pyenv/versions/3.5.7/lib/python3.5/site-packages/pandas/core/frame.py:7138: FutureWarning: Sorting because non-concatenation axis is not aligned. A future version
of pandas will change to not sort by default.
To accept the future behavior, pass 'sort=False'.
To retain the current behavior and silence the warning, pass 'sort=True'.
sort=sort,
| 0 | a | b | d | note | |
|---|---|---|---|---|---|
| 0 | 2.0 | 0 | 2.0 | 2.0 | NaN |
| 1 | 2.0 | 0 | 0.0 | 2.0 | NaN |
| 2 | 2.0 | 0 | 0.0 | 2.0 | NaN |
| 0 | NaN | 5 | NaN | NaN | 2.0 |
| 1 | NaN | 2 | 4.0 | NaN | 2.0 |
| 2 | NaN | -2 | -15.0 | NaN | 2.0 |
Merging DataFrames
df_account = pd.DataFrame({'accountNumber': ["AC1", "AC2", "AC3", "AC4"],
'Amount': [10000, 109300, 2984, 1999],
'Name': ["LIVRET A", "Compte Épargne Retraite", "Quadretto", "Compte Courant"]})
df_client = pd.DataFrame({'id_account': ["AC1", "AC2", "AC3", "AC4", "AC5"],
'Name': ["Luc", "René", "Jean", "Jean", "Joseph"],
'id_client': ["ID1099", "ID1091", "ID1018", "ID1018", "ID1021"]})
df_account
df_client
| Amount | Name | accountNumber | |
|---|---|---|---|
| 0 | 10000 | LIVRET A | AC1 |
| 1 | 109300 | Compte Épargne Retraite | AC2 |
| 2 | 2984 | Quadretto | AC3 |
| 3 | 1999 | Compte Courant | AC4 |
| Name | id_account | id_client | |
|---|---|---|---|
| 0 | Luc | AC1 | ID1099 |
| 1 | René | AC2 | ID1091 |
| 2 | Jean | AC3 | ID1018 |
| 3 | Jean | AC4 | ID1018 |
| 4 | Joseph | AC5 | ID1021 |
pd.merge(left=df_account, right=df_client,
left_on="accountNumber",
right_on="id_account",
how='inner')
| Amount | Name_x | accountNumber | Name_y | id_account | id_client | |
|---|---|---|---|---|---|---|
| 0 | 10000 | LIVRET A | AC1 | Luc | AC1 | ID1099 |
| 1 | 109300 | Compte Épargne Retraite | AC2 | René | AC2 | ID1091 |
| 2 | 2984 | Quadretto | AC3 | Jean | AC3 | ID1018 |
| 3 | 1999 | Compte Courant | AC4 | Jean | AC4 | ID1018 |
df_merged = pd.merge(left=df_account, right=df_client,
left_on="accountNumber",
right_on="id_account",
how='right',
suffixes=["_account", "_client"])
df_merged
| Amount | Name_account | accountNumber | Name_client | id_account | id_client | |
|---|---|---|---|---|---|---|
| 0 | 10000.0 | LIVRET A | AC1 | Luc | AC1 | ID1099 |
| 1 | 109300.0 | Compte Épargne Retraite | AC2 | René | AC2 | ID1091 |
| 2 | 2984.0 | Quadretto | AC3 | Jean | AC3 | ID1018 |
| 3 | 1999.0 | Compte Courant | AC4 | Jean | AC4 | ID1018 |
| 4 | NaN | NaN | NaN | Joseph | AC5 | ID1021 |
# to drop the (same) column we have been merging on
df_merged.drop('id_account', axis=1)
| Amount | Name_account | accountNumber | Name_client | id_client | |
|---|---|---|---|---|---|
| 0 | 10000.0 | LIVRET A | AC1 | Luc | ID1099 |
| 1 | 109300.0 | Compte Épargne Retraite | AC2 | René | ID1091 |
| 2 | 2984.0 | Quadretto | AC3 | Jean | ID1018 |
| 3 | 1999.0 | Compte Courant | AC4 | Jean | ID1018 |
| 4 | NaN | NaN | NaN | Joseph | ID1021 |
#df_notes["eleve"] = (df_notes
# .eleve
# .astype("category")
# .cat.rename_categories(
# new_categories=
# ["eleve{}".format(i) for i in range(df_notes.eleve.nunique())]
# )
#)
Apply
For the following sections, we are going to use real world data of students’grades from an exam I gave 😜 The data has been anonymised to fit GDPR regulation.
It has been retrieved by scrapping automatically the online web app that stores the results from each passed quizz.
You will see along the way we will need to make multiple modifications to our original data.
# !curl --help
# option : -o, --output <file>
# Write to file instead of stdout
!curl https://raw.githubusercontent.com/Luc-Bertin/TDs_ESILV/master/td3_discover_pandas/notes_eleves.csv -o "notes.csv"
% Total % Received % Xferd Average Speed Time Time Time Current
Dload Upload Total Spent Left Speed
100 21979 100 21979 0 0 66805 0 --:--:-- --:--:-- --:--:-- 66805
df_notes = pd.read_csv("notes.csv", index_col=0)
# showing just the first n rows
df_notes.head(5)
# or the last n rows
df_notes.tail(5)
| eleve | note | groupe | quizz | |
|---|---|---|---|---|
| 0 | eleve0 | 71,43 % | Unknown | td1 |
| 1 | eleve1 | 100 % | Unknown | td1 |
| 2 | eleve4 | 71,43 % | Unknown | td1 |
| 3 | eleve6 | 42,86 % | Unknown | td1 |
| 4 | eleve8 | 57,14 % | Unknown | td1 |
| eleve | note | groupe | quizz | |
|---|---|---|---|---|
| 741 | eleve174 | 100 % | ibo5 | td3 |
| 742 | eleve166 | 66,67 % | ibo5 | td3 |
| 743 | eleve176 | 83,33 % | ibo5 | td3 |
| 744 | eleve186 | 100 % | ibo5 | td3 |
| 745 | eleve196 | 66,67 % | ibo5 | td3 |
On a Series object
Let’s define a function to be applied for each element of a column.
def function(val):
"""A fonction to be applied on each element of a pandas DataFrame column / Series """
# we need to return a value for each element computation
return val.upper()
df_notes.eleve.apply(function) # the function applies on each value in the column
0 ELEVE0
1 ELEVE1
2 ELEVE4
3 ELEVE6
4 ELEVE8
...
741 ELEVE174
742 ELEVE166
743 ELEVE176
744 ELEVE186
745 ELEVE196
Name: eleve, Length: 746, dtype: object
behind, apply is looping on each element of the column eleve and returning a value for each of them
on a DataFrame object
We can also use the apply method on a DataFrame object, but we need to provide an axis.
applied function won’t be fed a single column element this time but a Series.
a Series whose index depends on the axis we choose !
df_notes
| eleve | note | groupe | quizz | |
|---|---|---|---|---|
| 0 | eleve0 | 71,43 % | Unknown | td1 |
| 1 | eleve1 | 100 % | Unknown | td1 |
| 2 | eleve4 | 71,43 % | Unknown | td1 |
| 3 | eleve6 | 42,86 % | Unknown | td1 |
| 4 | eleve8 | 57,14 % | Unknown | td1 |
| ... | ... | ... | ... | ... |
| 741 | eleve174 | 100 % | ibo5 | td3 |
| 742 | eleve166 | 66,67 % | ibo5 | td3 |
| 743 | eleve176 | 83,33 % | ibo5 | td3 |
| 744 | eleve186 | 100 % | ibo5 | td3 |
| 745 | eleve196 | 66,67 % | ibo5 | td3 |
746 rows × 4 columns
def function(row):
"""A fonction to be applied on each row of the DataFrame
i.e. each row is indeed a pandas.Series object
passed-in the applied function at each loop iteration.
We will need later on to use axis=1 for Series to be the rows"""
# having a full row we can do many things to create
if int( row["eleve"][-1] ) % 2 == 0:
return "pair"
return "impair"
df_notes.apply(function, axis=1)
0 pair
1 impair
2 pair
3 pair
4 pair
...
741 pair
742 pair
743 pair
744 pair
745 pair
Length: 746, dtype: object
def function(row):
"""Another function on rows but returning a pandas.Series each time
i.e. then the final result will be a stack of pandas.Series along an axis or the other
i.e. <=> a DataFrame"""
if int( row["eleve"][-1] ) % 2 == 0:
odd_or_even = "pair"
odd_or_even = "impair"
cut_note = row["note"].split(',')[0] # what precedes the comma
return pd.Series([ odd_or_even, cut_note], index=['odd_or_even', 'cut_note'])
df_notes.apply(function, axis=1) # a pandas.Series for each row
| odd_or_even | cut_note | |
|---|---|---|
| 0 | impair | 71 |
| 1 | impair | 100 % |
| 2 | impair | 71 |
| 3 | impair | 42 |
| 4 | impair | 57 |
| ... | ... | ... |
| 741 | impair | 100 % |
| 742 | impair | 66 |
| 743 | impair | 83 |
| 744 | impair | 100 % |
| 745 | impair | 66 |
746 rows × 2 columns
To avoid having to define a loop we should use vectorized functions (we will talk about it later on). On integers (not the case here) use of vectorized functions can greatly improve computational speed. (C-loop)
def function(col):
"""Another function on cols this time"""
try:
return sum([int(x[:1]) for x in col])
except:
return "can't sum on this col"
#return pd.Series([ odd_or_even, cut_note], index=['odd_or_even', 'cut_note'])
df_notes.apply(function, axis=0) # a pandas.Series for each col
eleve can't sum on this col
note 3053
groupe can't sum on this col
quizz can't sum on this col
dtype: object
df_notes.note.str[:1].astype(float).sum()
3053.0
Manipulating columns with strings
Back to the definition of a vectorized function: it is a function that applies on the whole sequence rather than each element as input.
This is the case for numpy functions like np.mean, np.sum, np.std which apply on a numerically valued input array as a whole, so the loop is moved from the Python-level to the C one
Numeric types include:
int,float,datetime,bool,category. They excludeobjectdtype and can be held in contiguous memory blocks. See here too, concerning C contiguous array stored in memory when creating a numpy array.
Why are numpy operations more efficient than simple crude Python ? as we’ve seen earlier Everything in Python is an object. This includes, unlike C, numbers. Python types therefore have an overhead which does not exist with native C types. NumPy methods are usually C-based.
check here
np.vectorize is fake vectorisation. According to documentation: The vectorize function is provided primarily for convenience, not for performance. The implementation is essentially a for loop. It means there is no reazon in vectorize of function wich could be applied directly as it is in your example. Actually this could lead to degraded performance. Main goal of the “vectorize” is to hide a for loop from you code. But it will not avoid it neither change expected results.
This link provides a good an example of simple vectorization.
Numpy does not provide vectorization functions for arrays of strings.
Pandas provide vectorized str operations. Pros are that you don’t have to write any loop and can take the column/Series as a whole. Cons are that they are not actually faster than using a simply apply. String operations are inherently difficult to vectorize. Pandas treats strings as objects, and all operations on objects fall back to a slow, loopy implementation.
Already provided Pandas vectorized string methods available in .str.
df_notes["eleve"] = df_notes.eleve.str.capitalize()
df_notes
| eleve | note | groupe | quizz | |
|---|---|---|---|---|
| 0 | Eleve0 | 71,43 % | Unknown | td1 |
| 1 | Eleve1 | 100 % | Unknown | td1 |
| 2 | Eleve4 | 71,43 % | Unknown | td1 |
| 3 | Eleve6 | 42,86 % | Unknown | td1 |
| 4 | Eleve8 | 57,14 % | Unknown | td1 |
| ... | ... | ... | ... | ... |
| 741 | Eleve174 | 100 % | ibo5 | td3 |
| 742 | Eleve166 | 66,67 % | ibo5 | td3 |
| 743 | Eleve176 | 83,33 % | ibo5 | td3 |
| 744 | Eleve186 | 100 % | ibo5 | td3 |
| 745 | Eleve196 | 66,67 % | ibo5 | td3 |
746 rows × 4 columns
mask = df_notes.groupe.str.startswith("U")
mask
0 True
1 True
2 True
3 True
4 True
...
741 False
742 False
743 False
744 False
745 False
Name: groupe, Length: 746, dtype: bool
df_notes[mask]
| eleve | note | groupe | quizz | |
|---|---|---|---|---|
| 0 | Eleve0 | 71,43 % | Unknown | td1 |
| 1 | Eleve1 | 100 % | Unknown | td1 |
| 2 | Eleve4 | 71,43 % | Unknown | td1 |
| 3 | Eleve6 | 42,86 % | Unknown | td1 |
| 4 | Eleve8 | 57,14 % | Unknown | td1 |
| ... | ... | ... | ... | ... |
| 87 | Eleve202 | 57,14 % | Unknown | td1 |
| 88 | Eleve203 | 57,14 % | Unknown | td1 |
| 89 | Eleve204 | 71,43 % | Unknown | td1 |
| 90 | Eleve205 | 42,86 % | Unknown | td1 |
| 91 | Eleve207 | 42,86 % | Unknown | td1 |
92 rows × 4 columns
df_notes.note.str.split(',')
0 [71, 43 %]
1 [100 %]
2 [71, 43 %]
3 [42, 86 %]
4 [57, 14 %]
...
741 [100 %]
742 [66, 67 %]
743 [83, 33 %]
744 [100 %]
745 [66, 67 %]
Name: note, Length: 746, dtype: object
(df_notes.note
.str.replace("%","") # replace all occurences of "%" as ""
.str.replace(",", ".") # replace all occurences of "," as "."
.astype(float)
)
0 71.43
1 100.00
2 71.43
3 42.86
4 57.14
...
741 100.00
742 66.67
743 83.33
744 100.00
745 66.67
Name: note, Length: 746, dtype: float64
(df_notes.note
.str.findall("(\d+),?(\d+)?") #regex to find all matching groups in each element of the Series
.str[0] # vectorized element access in the column, works for all iterable, hence even a list in a pd.Series,
.str.join(".") # join the lists with "." rather than ','
.str.rstrip('.') # take off the last dot if exists
.astype(float) # convert to float type
)
0 71.43
1 100.00
2 71.43
3 42.86
4 57.14
...
741 100.00
742 66.67
743 83.33
744 100.00
745 66.67
Name: note, Length: 746, dtype: float64
serie_notes =\
(
df_notes.note
.str.extract("(\d+),?(\d+)?") # expand to multiple cols
.fillna(0) # fill NaN as 0 when no matched group
.astype(float) # convert to float
)
serie_notes[0] += serie_notes[1]/100
serie_notes.drop(1, axis=1,inplace=True)
df_notes.note = serie_notes
df_notes
| eleve | note | groupe | quizz | |
|---|---|---|---|---|
| 0 | Eleve0 | 71.43 | Unknown | td1 |
| 1 | Eleve1 | 100.00 | Unknown | td1 |
| 2 | Eleve4 | 71.43 | Unknown | td1 |
| 3 | Eleve6 | 42.86 | Unknown | td1 |
| 4 | Eleve8 | 57.14 | Unknown | td1 |
| ... | ... | ... | ... | ... |
| 741 | Eleve174 | 100.00 | ibo5 | td3 |
| 742 | Eleve166 | 66.67 | ibo5 | td3 |
| 743 | Eleve176 | 83.33 | ibo5 | td3 |
| 744 | Eleve186 | 100.00 | ibo5 | td3 |
| 745 | Eleve196 | 66.67 | ibo5 | td3 |
746 rows × 4 columns
Other interesting functions to mention
To compute the counts of unique values use : pd.Series.value_counts()
df_notes.groupe.value_counts(ascending=False)
ibo1 117
ibo5 115
ibo7 114
Unknown 92
ibo6 85
ibo3 85
ibo4 81
ibo2 57
Name: groupe, dtype: int64
To do a binning : i.e. group a number of more or less continuous values into a smaller number of “bins”. Use `pd.cut
pd.cut(df_notes.note, bins=5) # 5 equal sized bins
0 (60.0, 80.0]
1 (80.0, 100.0]
2 (60.0, 80.0]
3 (40.0, 60.0]
4 (40.0, 60.0]
...
741 (80.0, 100.0]
742 (60.0, 80.0]
743 (80.0, 100.0]
744 (80.0, 100.0]
745 (60.0, 80.0]
Name: note, Length: 746, dtype: category
Categories (5, interval[float64]): [(-0.1, 20.0] < (20.0, 40.0] < (40.0, 60.0] < (60.0, 80.0] < (80.0, 100.0]]
pd.cut(df_notes.note, bins=[0, 50, 75, 100])
0 (50, 75]
1 (75, 100]
2 (50, 75]
3 (0, 50]
4 (50, 75]
...
741 (75, 100]
742 (50, 75]
743 (75, 100]
744 (75, 100]
745 (50, 75]
Name: note, Length: 746, dtype: category
Categories (3, interval[int64]): [(0, 50] < (50, 75] < (75, 100]]
try:
pd.cut(df_notes.note, bins=[0, 50, 75, 100], labels=["Bad"])
except Exception as e:
print(e)
Bin labels must be one fewer than the number of bin edges
df_notes['appreciation'] = pd.cut(df_notes.note, bins=[0, 25, 50, 75, 100], labels=["Very Bad", "Bad", "Ok", "Good"])
df_notes.appreciation
0 Ok
1 Good
2 Ok
3 Bad
4 Ok
...
741 Good
742 Ok
743 Good
744 Good
745 Ok
Name: note, Length: 746, dtype: category
Categories (4, object): [Very Bad < Bad < Ok < Good]
GroupBy !
df_notes.head(3)
| eleve | note | groupe | quizz | |
|---|---|---|---|---|
| 0 | Eleve0 | 71.43 | Unknown | td1 |
| 1 | Eleve1 | 100.00 | Unknown | td1 |
| 2 | Eleve4 | 71.43 | Unknown | td1 |
Groupby applies the “split, apply, combine” method.
- We first have to use a
keyto groupby, i.e. a column of different labels that will serve to split the maindfinto different subsets (one for each label in the concerned column), just as we would do a GROUP BY in SQL syntax.
df_notes.groupby('groupe')
<pandas.core.groupby.generic.DataFrameGroupBy object at 0x1187d9a90>
No computation is done yet
This result in a DataFrameGroupBy object we can iterate on.
for name_group, group in df_notes.groupby('groupe'):
# the label used , the df subset (one for each label)
print( "label used {}, dataframe shape {}".format(name_group,group.shape))
label used Unknown, dataframe shape (92, 4)
label used ibo1, dataframe shape (117, 4)
label used ibo2, dataframe shape (57, 4)
label used ibo3, dataframe shape (85, 4)
label used ibo4, dataframe shape (81, 4)
label used ibo5, dataframe shape (115, 4)
label used ibo6, dataframe shape (85, 4)
label used ibo7, dataframe shape (114, 4)
- Notice that we are not limited by grouping over one column keys.
for name_group, group in df_notes.groupby(['groupe', "quizz"]):
# the label used , the df subset (one for each label)
print( "label used {}, dataframe shape {}".format(name_group,group.shape))
label used ('Unknown', 'td1'), dataframe shape (92, 4)
label used ('ibo1', 'td1'), dataframe shape (30, 4)
label used ('ibo1', 'td2'), dataframe shape (30, 4)
label used ('ibo1', 'td3'), dataframe shape (27, 4)
label used ('ibo1', 'td4'), dataframe shape (30, 4)
label used ('ibo2', 'td2'), dataframe shape (30, 4)
label used ('ibo2', 'td3'), dataframe shape (27, 4)
label used ('ibo3', 'td2'), dataframe shape (30, 4)
label used ('ibo3', 'td3'), dataframe shape (27, 4)
label used ('ibo3', 'td4'), dataframe shape (28, 4)
label used ('ibo4', 'td2'), dataframe shape (27, 4)
label used ('ibo4', 'td3'), dataframe shape (28, 4)
label used ('ibo4', 'td4'), dataframe shape (26, 4)
label used ('ibo5', 'td1'), dataframe shape (27, 4)
label used ('ibo5', 'td2'), dataframe shape (30, 4)
label used ('ibo5', 'td3'), dataframe shape (28, 4)
label used ('ibo5', 'td4'), dataframe shape (30, 4)
label used ('ibo6', 'td2'), dataframe shape (29, 4)
label used ('ibo6', 'td3'), dataframe shape (28, 4)
label used ('ibo6', 'td4'), dataframe shape (28, 4)
label used ('ibo7', 'td1'), dataframe shape (29, 4)
label used ('ibo7', 'td2'), dataframe shape (28, 4)
label used ('ibo7', 'td3'), dataframe shape (28, 4)
label used ('ibo7', 'td4'), dataframe shape (29, 4)
This results in a mutli-index with:
- level0 = the group
- level1 = the quizz number
We can also index the GroupByDataFrame object by retrieving one Series (again no computation is done yet)
df_notes.groupby(['groupe', "quizz"])["note"]
<pandas.core.groupby.generic.SeriesGroupBy object at 0x1188bdeb8>
for name_group, group in df_notes.groupby(['groupe', "quizz"])["note"]:
print( "label used {}, \n{} shape {}".format(name_group, type(group), group.shape))
label used ('Unknown', 'td1'),
<class 'pandas.core.series.Series'> shape (92,)
label used ('ibo1', 'td1'),
<class 'pandas.core.series.Series'> shape (30,)
label used ('ibo1', 'td2'),
<class 'pandas.core.series.Series'> shape (30,)
label used ('ibo1', 'td3'),
<class 'pandas.core.series.Series'> shape (27,)
label used ('ibo1', 'td4'),
<class 'pandas.core.series.Series'> shape (30,)
label used ('ibo2', 'td2'),
<class 'pandas.core.series.Series'> shape (30,)
label used ('ibo2', 'td3'),
<class 'pandas.core.series.Series'> shape (27,)
label used ('ibo3', 'td2'),
<class 'pandas.core.series.Series'> shape (30,)
label used ('ibo3', 'td3'),
<class 'pandas.core.series.Series'> shape (27,)
label used ('ibo3', 'td4'),
<class 'pandas.core.series.Series'> shape (28,)
label used ('ibo4', 'td2'),
<class 'pandas.core.series.Series'> shape (27,)
label used ('ibo4', 'td3'),
<class 'pandas.core.series.Series'> shape (28,)
label used ('ibo4', 'td4'),
<class 'pandas.core.series.Series'> shape (26,)
label used ('ibo5', 'td1'),
<class 'pandas.core.series.Series'> shape (27,)
label used ('ibo5', 'td2'),
<class 'pandas.core.series.Series'> shape (30,)
label used ('ibo5', 'td3'),
<class 'pandas.core.series.Series'> shape (28,)
label used ('ibo5', 'td4'),
<class 'pandas.core.series.Series'> shape (30,)
label used ('ibo6', 'td2'),
<class 'pandas.core.series.Series'> shape (29,)
label used ('ibo6', 'td3'),
<class 'pandas.core.series.Series'> shape (28,)
label used ('ibo6', 'td4'),
<class 'pandas.core.series.Series'> shape (28,)
label used ('ibo7', 'td1'),
<class 'pandas.core.series.Series'> shape (29,)
label used ('ibo7', 'td2'),
<class 'pandas.core.series.Series'> shape (28,)
label used ('ibo7', 'td3'),
<class 'pandas.core.series.Series'> shape (28,)
label used ('ibo7', 'td4'),
<class 'pandas.core.series.Series'> shape (29,)
Aggregation functions
We can now think about the “apply, combine” part
df_notes.dtypes
eleve object
note float64
groupe object
quizz object
dtype: object
Pandas provides us some functions to be applied on a dataframe or Series (.mean(), .sum(), .std(), .describe(), .min(), etc…), we can seemlessly append one of them to the GroupBy Object to operate on each of the subsets DataFrames/Series created on the split step (this is the apply step).
After applying the function to each split, a combined result is returned, in the form of a Series object or DataFrame.
Note that for those aggregating functions reduce the shape of the data e.g. summing or meaning on a Series result in a scalar (the sum or the mean), this will be operated over each Series groups from the split step.
df_notes.groupby(['groupe'])["note"].mean()
groupe
Unknown 63.664022
ibo1 87.337607
ibo2 97.251053
ibo3 86.418824
ibo4 87.953580
ibo5 80.288957
ibo6 83.484000
ibo7 86.402456
Name: note, dtype: float64
df_notes.groupby(['groupe', 'quizz'])["note"].mean()
groupe quizz
Unknown td1 63.664022
ibo1 td1 67.618000
td2 94.666667
td3 90.739630
td4 96.666333
ibo2 td2 98.666667
td3 95.678148
ibo3 td2 88.666667
td3 88.887037
td4 81.630357
ibo4 td2 87.407407
td3 87.500000
td4 89.009231
ibo5 td1 53.967407
td2 90.000000
td3 90.475714
td4 84.759667
ibo6 td2 90.344828
td3 83.332857
td4 76.529286
ibo7 td1 74.875517
td2 85.714286
td3 94.642500
td4 90.637931
Name: note, dtype: float64
as we get a hierarchical index we can unstack to make use of the dimensionality brought by column indexesm
_.unstack()
| quizz | td1 | td2 | td3 | td4 |
|---|---|---|---|---|
| groupe | ||||
| Unknown | 63.664022 | NaN | NaN | NaN |
| ibo1 | 67.618000 | 94.666667 | 90.739630 | 96.666333 |
| ibo2 | NaN | 98.666667 | 95.678148 | NaN |
| ibo3 | NaN | 88.666667 | 88.887037 | 81.630357 |
| ibo4 | NaN | 87.407407 | 87.500000 | 89.009231 |
| ibo5 | 53.967407 | 90.000000 | 90.475714 | 84.759667 |
| ibo6 | NaN | 90.344828 | 83.332857 | 76.529286 |
| ibo7 | 74.875517 | 85.714286 | 94.642500 | 90.637931 |
Something is unusual? why is there NaN? some class groups should have grades for each quizz.
df_notes.isnull().sum()
eleve 0
note 0
groupe 0
quizz 0
dtype: int64
though all the data seems complete…
df_notes.isnull().apply(sum, axis=0)
eleve 0
note 0
groupe 0
quizz 0
dtype: int64
Notice the Unknown group, we should look more into this…
df_notes[df_notes.groupe == "Unknown"]
| eleve | note | groupe | quizz | appreciation | |
|---|---|---|---|---|---|
| 0 | Eleve0 | 71.43 | Unknown | td1 | Ok |
| 1 | Eleve1 | 100.00 | Unknown | td1 | Good |
| 2 | Eleve4 | 71.43 | Unknown | td1 | Ok |
| 3 | Eleve6 | 42.86 | Unknown | td1 | Bad |
| 4 | Eleve8 | 57.14 | Unknown | td1 | Ok |
| ... | ... | ... | ... | ... | ... |
| 87 | Eleve202 | 57.14 | Unknown | td1 | Ok |
| 88 | Eleve203 | 57.14 | Unknown | td1 | Ok |
| 89 | Eleve204 | 71.43 | Unknown | td1 | Ok |
| 90 | Eleve205 | 42.86 | Unknown | td1 | Bad |
| 91 | Eleve207 | 42.86 | Unknown | td1 | Bad |
92 rows × 5 columns
_.shape[0]
92
To apply multiple aggregate functions at once using a list of the functions you want to apply in aggregate
df_notes.groupby('quizz').agg({'note': ['max', min]})
| note | ||
|---|---|---|
| max | min | |
| quizz | ||
| td1 | 100.0 | 0.00 |
| td2 | 100.0 | 40.00 |
| td3 | 100.0 | 50.00 |
| td4 | 100.0 | 42.86 |
this results in a multi column index
_.columns
MultiIndex([('note', 'max'),
('note', 'min')],
)
Grouping by students, we may have more insight.
df_notes.groupby('eleve').agg(list)
# applied on all columns (where the function can be used on) for each subset
| note | groupe | quizz | |
|---|---|---|---|
| eleve | |||
| Eleve0 | [71.43, 80.0, 100.0] | [Unknown, ibo2, ibo2] | [td1, td2, td3] |
| Eleve1 | [100.0, 100.0, 100.0] | [Unknown, ibo2, ibo2] | [td1, td2, td3] |
| Eleve10 | [100.0, 100.0, 85.71, 71.43] | [ibo7, ibo7, ibo7, ibo7] | [td3, td2, td4, td1] |
| Eleve100 | [100.0, 100.0, 100.0, 85.71] | [ibo7, ibo7, ibo7, ibo7] | [td3, td2, td4, td1] |
| Eleve101 | [100.0, 100.0, 85.71, 100.0] | [ibo7, ibo7, ibo7, ibo7] | [td3, td2, td4, td1] |
| ... | ... | ... | ... |
| Eleve95 | [85.71, 100.0, 100.0] | [Unknown, ibo2, ibo2] | [td1, td2, td3] |
| Eleve96 | [100.0, 100.0, 100.0, 42.86] | [ibo1, ibo1, ibo1, ibo1] | [td4, td3, td2, td1] |
| Eleve97 | [28.57, 85.71, 83.33, 60.0] | [Unknown, ibo4, ibo4, ibo4] | [td1, td4, td3, td2] |
| Eleve98 | [42.86, 85.71, 100.0, 100.0] | [Unknown, ibo4, ibo4, ibo4] | [td1, td4, td3, td2] |
| Eleve99 | [71.43, 100.0, 100.0] | [Unknown, ibo2, ibo2] | [td1, td2, td3] |
208 rows × 3 columns
Some students are known, but are not always written as such.
transform
sometimes we want the “apply-combine” steps to avoid reducing the data size but compute for each data record something based on some intra-group/splits caracteristics
Here we would want for example to group by students and replace Unknown fields by the correct information from the other students record.
df_notes
| eleve | note | groupe | quizz | |
|---|---|---|---|---|
| 0 | Eleve0 | 71.43 | ibo2 | td1 |
| 1 | Eleve1 | 100.00 | ibo2 | td1 |
| 2 | Eleve4 | 71.43 | ibo2 | td1 |
| 3 | Eleve6 | 42.86 | ibo6 | td1 |
| 4 | Eleve8 | 57.14 | ibo4 | td1 |
| ... | ... | ... | ... | ... |
| 741 | Eleve174 | 100.00 | ibo5 | td3 |
| 742 | Eleve166 | 66.67 | ibo5 | td3 |
| 743 | Eleve176 | 83.33 | ibo5 | td3 |
| 744 | Eleve186 | 100.00 | ibo5 | td3 |
| 745 | Eleve196 | 66.67 | ibo5 | td3 |
746 rows × 4 columns
df_notes.replace({"Unknown":np.nan}, inplace=True)
df_notes["groupe"] = df_notes.groupby('eleve')['groupe'].transform(lambda x: x.bfill().ffill())
df_notes.groupby('eleve').agg(list)
| note | groupe | quizz | appreciation | |
|---|---|---|---|---|
| eleve | ||||
| Eleve0 | [71.43, 80.0, 100.0] | [ibo2, ibo2, ibo2] | [td1, td2, td3] | [Ok, Good, Good] |
| Eleve1 | [100.0, 100.0, 100.0] | [ibo2, ibo2, ibo2] | [td1, td2, td3] | [Good, Good, Good] |
| Eleve10 | [100.0, 100.0, 85.71, 71.43] | [ibo7, ibo7, ibo7, ibo7] | [td3, td2, td4, td1] | [Good, Good, Good, Ok] |
| Eleve100 | [100.0, 100.0, 100.0, 85.71] | [ibo7, ibo7, ibo7, ibo7] | [td3, td2, td4, td1] | [Good, Good, Good, Good] |
| Eleve101 | [100.0, 100.0, 85.71, 100.0] | [ibo7, ibo7, ibo7, ibo7] | [td3, td2, td4, td1] | [Good, Good, Good, Good] |
| ... | ... | ... | ... | ... |
| Eleve95 | [85.71, 100.0, 100.0] | [ibo2, ibo2, ibo2] | [td1, td2, td3] | [Good, Good, Good] |
| Eleve96 | [100.0, 100.0, 100.0, 42.86] | [ibo1, ibo1, ibo1, ibo1] | [td4, td3, td2, td1] | [Good, Good, Good, Bad] |
| Eleve97 | [28.57, 85.71, 83.33, 60.0] | [ibo4, ibo4, ibo4, ibo4] | [td1, td4, td3, td2] | [Bad, Good, Good, Ok] |
| Eleve98 | [42.86, 85.71, 100.0, 100.0] | [ibo4, ibo4, ibo4, ibo4] | [td1, td4, td3, td2] | [Bad, Good, Good, Good] |
| Eleve99 | [71.43, 100.0, 100.0] | [ibo2, ibo2, ibo2] | [td1, td2, td3] | [Ok, Good, Good] |
208 rows × 4 columns
Let’s check if we still have some NAs?
df_notes.groupby(['groupe', 'quizz'])["note"].mean().unstack()
| quizz | td1 | td2 | td3 | td4 |
|---|---|---|---|---|
| groupe | ||||
| ibo1 | 67.618000 | 94.666667 | 90.739630 | 96.666333 |
| ibo2 | 76.846207 | 98.666667 | 95.678148 | NaN |
| ibo3 | 40.002000 | 88.666667 | 88.887037 | 81.630357 |
| ibo4 | 56.632500 | 87.407407 | 87.500000 | 89.009231 |
| ibo5 | 53.967407 | 90.000000 | 90.475714 | 84.759667 |
| ibo6 | 61.427667 | 90.344828 | 83.332857 | 76.529286 |
| ibo7 | 74.875517 | 85.714286 | 94.642500 | 90.637931 |
Seems better !
df_notes[(df_notes.groupe == 'ibo2') & (df_notes.quizz == 'td4')]
| eleve | note | groupe | quizz |
|---|
seems we don’t have data for the exam number 4 fro this group (which had been cancelled due too large manifestations in Paris which lead to postpone the session too late.)
df_notes = df_notes[~(df_notes.quizz=='td4')]
Pivot Table
df_notes.groupby(['groupe', 'quizz'])["note"].mean().unstack()
| quizz | td1 | td2 | td3 |
|---|---|---|---|
| groupe | |||
| ibo1 | 67.618000 | 94.666667 | 90.739630 |
| ibo2 | 76.846207 | 98.666667 | 95.678148 |
| ibo3 | 40.002000 | 88.666667 | 88.887037 |
| ibo4 | 56.632500 | 87.407407 | 87.500000 |
| ibo5 | 53.967407 | 90.000000 | 90.475714 |
| ibo6 | 61.427667 | 90.344828 | 83.332857 |
| ibo7 | 74.875517 | 85.714286 | 94.642500 |
df_notes.pivot_table('note', index='groupe', columns='quizz', margins=True)
| quizz | td1 | td2 | td3 | All |
|---|---|---|---|---|
| groupe | ||||
| ibo1 | 67.618000 | 94.666667 | 90.739630 | 84.120805 |
| ibo2 | 76.846207 | 98.666667 | 95.678148 | 90.370349 |
| ibo3 | 40.002000 | 88.666667 | 88.887037 | 84.838065 |
| ibo4 | 56.632500 | 87.407407 | 87.500000 | 77.056747 |
| ibo5 | 53.967407 | 90.000000 | 90.475714 | 78.711059 |
| ibo6 | 61.427667 | 90.344828 | 83.332857 | 78.116667 |
| ibo7 | 74.875517 | 85.714286 | 94.642500 | 84.957412 |
| All | 64.686180 | 90.882353 | 90.154715 | 82.528696 |
results = df_notes.pivot_table(index='groupe', columns='quizz',
aggfunc={"note":['max', min]})
results
| note | ||||||
|---|---|---|---|---|---|---|
| max | min | |||||
| quizz | td1 | td2 | td3 | td1 | td2 | td3 |
| groupe | ||||||
| ibo1 | 100.00 | 100.0 | 100.0 | 28.57 | 80.0 | 50.00 |
| ibo2 | 100.00 | 100.0 | 100.0 | 42.86 | 80.0 | 83.33 |
| ibo3 | 57.14 | 100.0 | 100.0 | 14.29 | 40.0 | 66.67 |
| ibo4 | 100.00 | 100.0 | 100.0 | 0.00 | 40.0 | 66.67 |
| ibo5 | 85.71 | 100.0 | 100.0 | 0.00 | 60.0 | 50.00 |
| ibo6 | 100.00 | 100.0 | 100.0 | 28.57 | 40.0 | 50.00 |
| ibo7 | 100.00 | 100.0 | 100.0 | 28.57 | 40.0 | 66.67 |
%matplotlib inline
import matplotlib.pyplot as plt
plt.rcParams['figure.figsize'] = (20, 10)
results['note'].plot(kind='bar')
<matplotlib.axes._subplots.AxesSubplot at 0x11d11eba8>
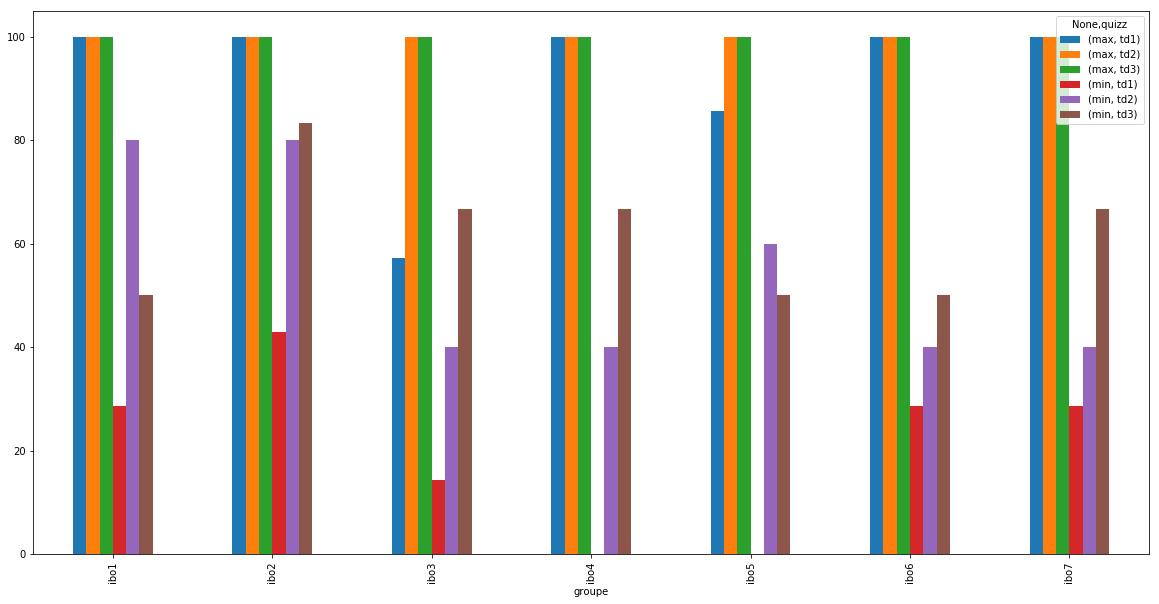
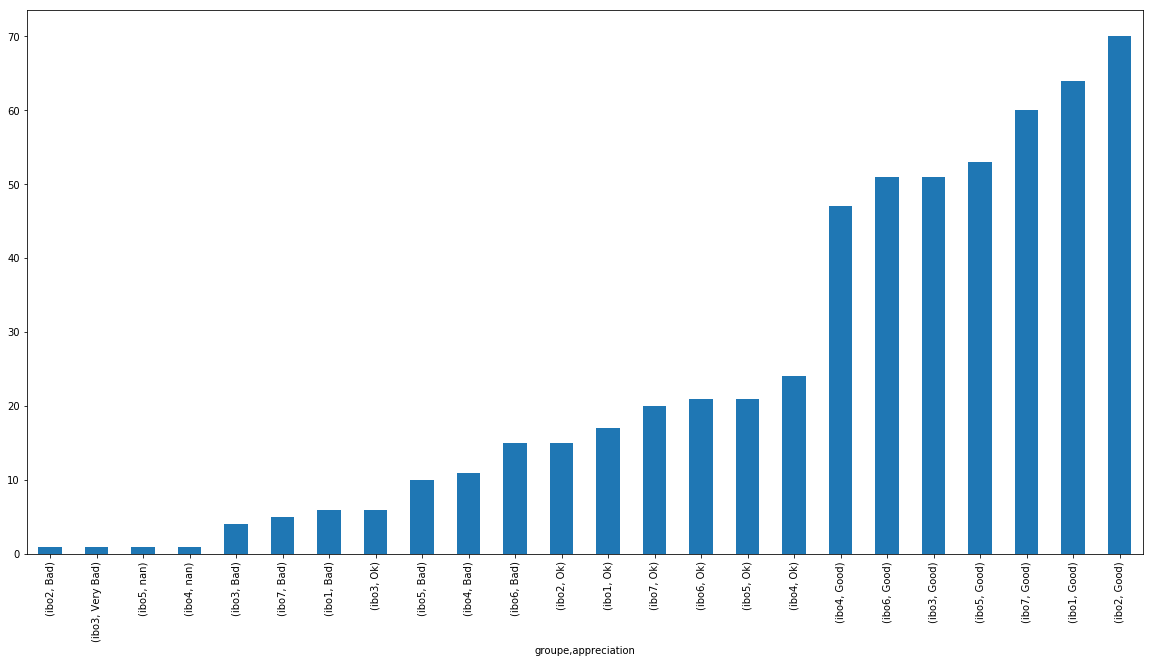
df_notes.groupby('groupe').appreciation.value_counts(dropna=False).sort_values().plot(kind="bar")
<matplotlib.axes._subplots.AxesSubplot at 0x11ddc1cc0>
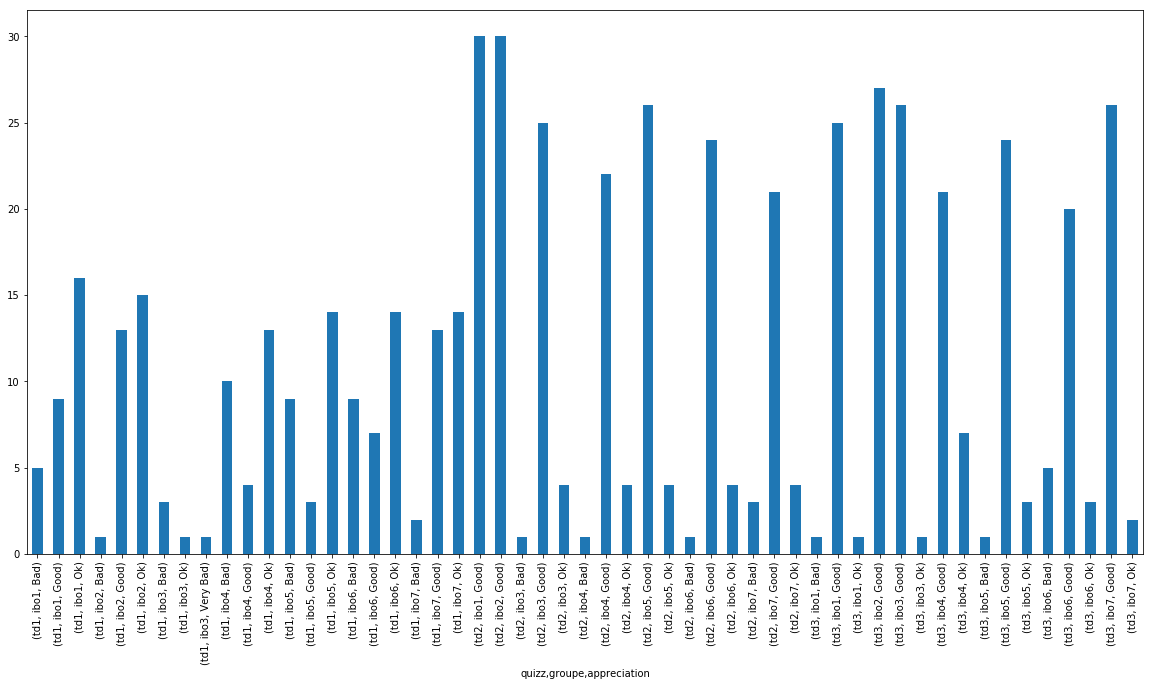
(df_notes.
.groupby(['quizz','groupe'])
['appreciation']
.value_counts().sort_index()
.plot(kind="bar"))
<matplotlib.axes._subplots.AxesSubplot at 0x11d1b4908>
(df_notes
.groupby(['quizz','groupe'])
['appreciation']
.value_counts()
.unstack().plot(kind='bar', stacked=True))
<matplotlib.axes._subplots.AxesSubplot at 0x11ef2ff98>
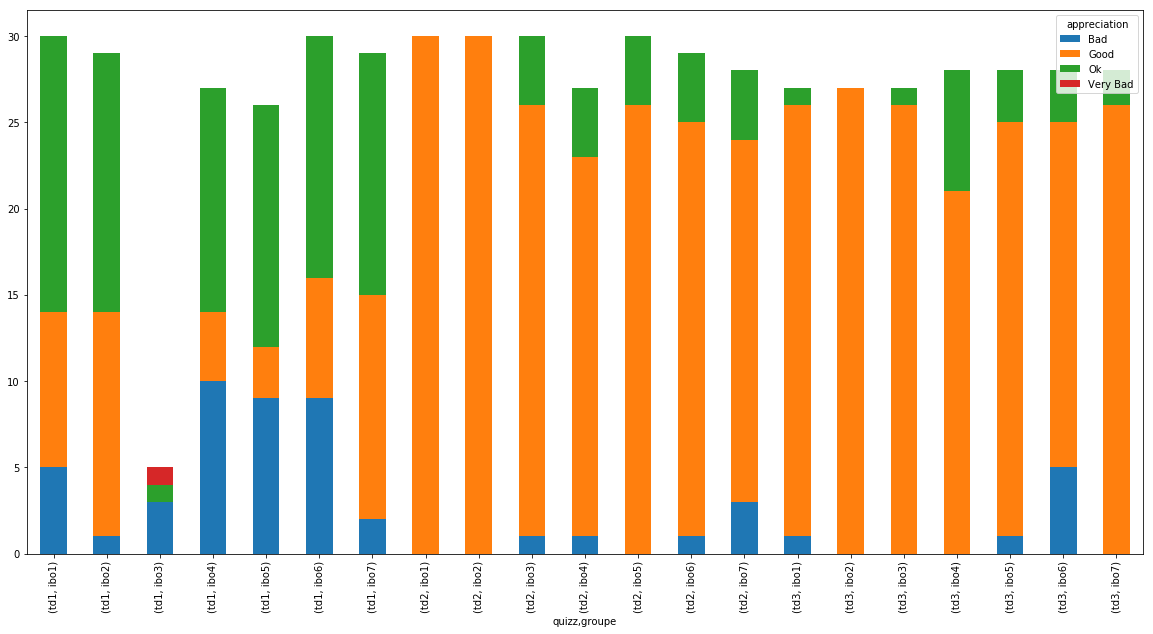
(df_notes
.pivot_table(index='quizz',
columns='groupe',
aggfunc={'appreciation':'value_counts'})
.plot(kind="bar"))
<matplotlib.axes._subplots.AxesSubplot at 0x11ee1d7f0>
df_notes.appreciation.value_counts(dropna=False).plot(kind="bar")
<matplotlib.axes._subplots.AxesSubplot at 0x11d5a9128>
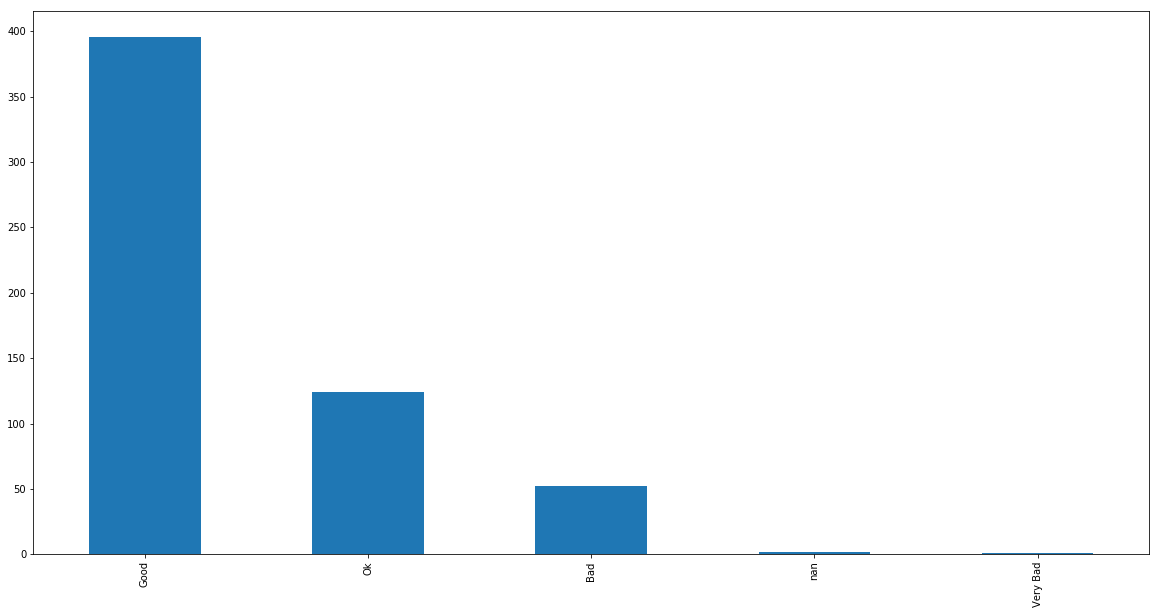
df_notes[pd.isna(df_notes.appreciation)]
| eleve | note | groupe | quizz | appreciation | |
|---|---|---|---|---|---|
| 75 | Eleve183 | 0.0 | ibo4 | td1 | NaN |
| 525 | Eleve111 | 0.0 | ibo5 | td1 | NaN |
missing grades for students.
Final question should be…
What should we put in fillna for them 😉 ?
Just an interesting link on representation of the data

 Webscrapping using Selenium
Webscrapping using Selenium